Abstract
Background:
Methicillin -resistant staphylococcus aureus (MRSA) is associated with serious infections. Having the ability of biofilm-formation decrease their susceptibility to antibiotics.Objectives:
The aim of this study was to determine the prevalence of biofilm formation among MRSA isolated from nasal carriers in the Beheshti Teaching Hospital in Kashan, Iran.Materials and Methods:
A cross-sectional study was conducted in 810 patients referred to emergency department in Beheshti Hospital in Kashan. Sterilized nasal swabs were used for collecting nasal bacteria. Nasal specimens were further recognized as S. aureus strains by standard biochemical tests, and MRSA isolates were detected by disk diffusion method. PCR assay was used for detecting mecA gene in MRSA isolates. The susceptibility of MRSA isolates to amikacin, clindamycin, gentamicin, ciprofloxacin, SXT, erythromycin, tetracycline were determined by using disk diffusion method according to recommendation of CLSI. Biofilm formation ability of MRSA isolates were examined by crystal violet microtitre plate assay and Congo red agar (CRA).Results:
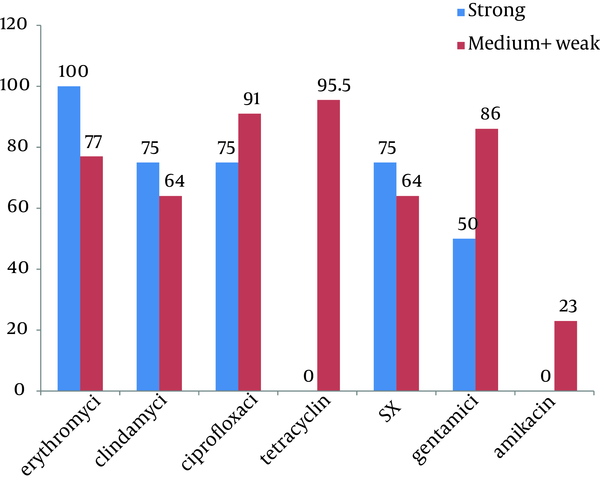
Two hundred and ninety six (36.5%) out of 810 isolates were S. aureus. Twenty six (8.8%) of all S. aureus isolates were recognized as MRSA. All the MRSA isolates have the ability of biofilm formation which 15.4%, 19.2% and 65.4% of them were strong, medium and weak biofilm producer respectively. The resistance rate of strong biofilm producer were; erythromycin (100%), clindamycin (75%), ciprofloxacin (75%), SXT (75%), gentamycin (50%), tetracycline (0%), amikacin (0%).Conclusions:
High rate of MRSA nasal carrier and having the ability of biofilm formation which decrease their susceptibility to antibiotics, is an alarming for public health. Statistically significant correlation between susceptibility to tetracycline and MRSA carrier was observed.Keywords
Methicillin-resistant Staphylococcus aureus Biofilm formation Antibiotic Susceptibility
1. Background
Staphylococcus aureus is known to form biofilms on different surfaces (1). The chronic infections that cause by S. aureus, persist and increase the rate of morbidity and mortality in human population due to the development of biofilm structures produced by this pathogen (2). Biofilm forming bacteria are the cause of many nosocomial infections (3). According to some reports, over 65% of hospital-acquired infections occur by the infecting organisms that have the ability of producing biofilms (4). Biofilms are the population of bacteria growing on the biotic and abiotic surfaces and embed themselves in a self-produced extracellular matrix of exopolysaccharide (EPS), proteins and some micro molecules such as DNA (2, 5). There are various definitions for biofilm but all of them enumerate three major ingredients for it: microbes, slime exopolysaccharide and surface, removing any of them can stop developing biofilm (6).
Susceptibility to antibiotics in bacteria that are protected by biofilm is reduced because drugs are prevented from reaching the bacteria surrounded by biofilm. Furthermore biofilm keeps bacteria out of reach of host immune defense mechanism and often resulting in persistent and difficult-to-treat infections (2, 3). Methicillin resistant S. aureus (MRSA) that have the ability of biofilm formation can become resistant to the most currently use antibiotics (1). MRSA infections are life-threatening due to emergence of multidrug resistance strains and also occurrence of isolates that are able to form strong biofilms (7). Early identification and adopting efficient control protocol against biofilm forming MRSA can be one of the essential steps towards the prevention of the most serious nosocomial infections.
2. Objectives
The aim of this study was to determine the prevalence of biofilm formation among MRSA isolated from nasal carriers referred to emergency department in the Beheshti Teaching Hospital in Kashan, Iran.
3. Materials and Methods
During the period November 2011 to Jun 2012, eight hundred and ten of people who aged over 18 years, referred to emergency department, Beheshti hospital, Kasahan, Iran participated in this study. A questionnaire including risk factors for MRSA nasal colonization was filled for each patient. Informed consent was obtained from all participants, and the study was approved by the ethics committees of Kashan University of Medical Sciences. Sterile swabs were used for collecting samples from both anterior nares.
3.1. Laboratory Methods
Samples were cultured on blood agar and incubated at 37ºC for 24 h. S. aureus isolates were confirmed by Gram staining, catalase, oxidase, coagulase and growth characteristics on mannitol-salt agar. MRSA isolates were detected by oxacillin (1 µg) and using disk diffusion method according to Clinical and Laboratory Standards Institute guidelines (8).
3.2. Antimicrobial Susceptibility Tests
Antibiotic susceptibility pattern to amikacin (30 µg), clindamycin (2 µg), gentamicin (10 µg), ciprofloxacin (5 µg), trimethoprim-sulfamethoxazole (1.25 / 23.75 µg), erythromycin (15 µg), tetracycline (30 µg) were determined by using disk diffusion method according to recommendation of Clinical and Laboratory Standards Institute (8).
3.3. Polymerase Chain Reaction Detection of mecA
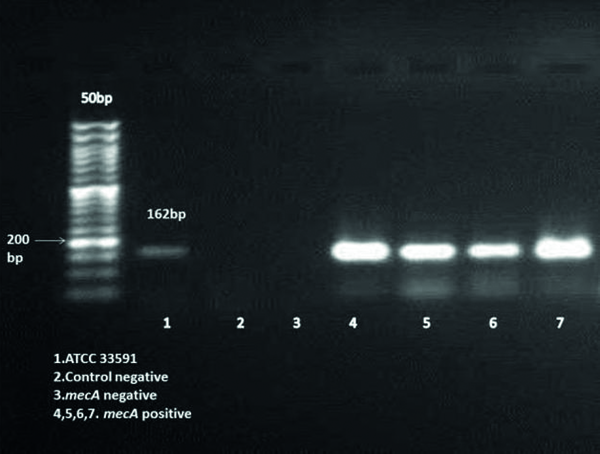
The presence of mecA gene in MRSA isolates was confirmed by a PCR assay. The following primers were used in this assay: Forward 5'TCCAGATTACAACTTCACCAGG3', Reverse 5'CCACTTCATATCTTGTAACG3'. These primers amplify 162 bp of DNA fragment (9). The reaction was carried out in a 25 µl volume containing: 2.5 µl of 10X PCR Buffer, 1.5 mM Mgcl2, 0.2 µM dNTP Mix, 0.5 µM of each primer, 5ng template DNA, 0.05 u/µl Taq DNA polymerase. Ultra pure water was then added to make up a final volume of 25 µl. polymerase chain reaction was carried out with thermal cycler (Mastercycler gradient; eppendorf, Germany).
The amplification cycles consisted of an initial denaturation of target DNA at 95°C for 15 min was followed by 30 cycles of initial denaturation at 94°C for 30s, 57°C for 1.5 min and 72°C for 1.5 min, ending with a final extension step at 72°C for 10 min, holding step at 4°C. In each PCR run, S. aureus strain ATCC 33591 was used as a positive control and negative controls were added to each PCR run. PCR products were seen by 1.8% agarose gel electrophoresis. A 50 bp DNA ladder was used as a size marker. The amplified bands were visualized under ultraviolet light and photographed.
3.3. Biofilm Formation Assay
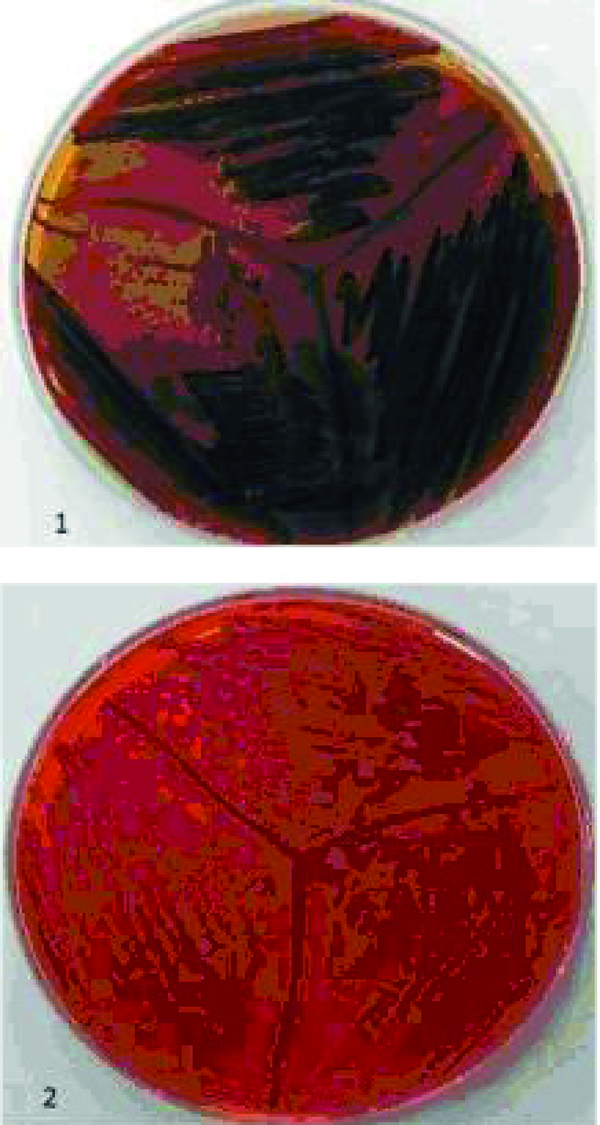
Congo red agar was used for detecting slime producing isolates. MRSA isolates were cultured on the agar containing 10g of glucose with 0.4 g of Congo red (Merck, Germany ) in one liter of Blood Base Agar-2 (BAB-2) and incubated at 37 ºC for 48 h. Strains which produced black colonies considered as slime producers and strains with red colonies labeled as non-slime producers (3). The ability of MRSA isolates to produce biofilm was determined by a method reported previously (10). In this method, MRSA strains were grown overnight at 37ºC in tryptic soy broth containing 0.25% glucose.
The culture was diluted 1:100 in medium. Sterile flat -bottomed 96-well poly styrene microtiter plates were inoculated with 200 µl of bacterial suspension, and incubated for 24 h at 37 ºC without agitation. Wells were washed three times with 300 µl of distilled water, dried in an inverted position at room temperature and finally stained with 300µl of 2% crystal violet solution in water for 45 min. After staining, wells were washed 3 times with distilled water. For destaining the wells 300µl of ethanol–acetic acid (95:5 vol/vol) were added to each well. A new sterile flat-bottomed 96-well poly styrene microtiter plates were inoculated with 100 µl destaining solution of each well. The absorbance of destaining solution was measured at 570 nm in an Elisa reader (Stat fax-2100). Each test was done triplicate. As a control, uninoculated medium was used. The mean OD570 value from control wells subtracted from the mean OD570 value of tested wells.
3.4. Statistical Analysis
SPSS software (SPSS Inc no.16) was used for data analysis. Fischer exact test or χ2-test was used for analysis of categorical data. A P value of < 0.05 was considered statistically significant.
4. Results
Two hundred and ninety six (36.5%) S. aureus were isolated from eight hundred and ten participants. Thirty two MRSA were detected by disk diffusion method but mecA gene was seen in twenty six of them. Figure 1 shows the amplification for presence of mecA gene. Distribution of MRSA nasal carrier according to age group, sex and ward are summarized in Table 1. All of 26 isolates were able to form biofilm in various levels. Four (15.4%), Five (19.2%) and 17 (65.4%) of MRSA isolates produced strong (OD570 0 to < 0.2), medium (OD570 ≥ 0.2 to < 0.5) and weak (OD570 ≥ 0.5) biofilm respectively. Slime layer assay showed that 18 0ut of 26 (69.2%) of MRSA isolates were slime layer producer and 8 0ut of 26 (30.8%) of them were classified as non-slime layer producer. Figure 2 shows colonies of slime layer and non-slime layer producer MRSA isolates on CRA.
Gel Electrophoresis of the PCR Amplification Using mecA Gene Specific Primers
Black Colonies (1) on CRA Show Slime Layer Producer MRSA and red Colonies (2) Illustrate non-Slime Layer Producer MRSA
Distribution of MRSA Nasal Carrier According to age Group, sex and Ward
| Parameter | No. (%) |
|---|---|
| Age, y | |
| 18 - 23 | 0 (0) |
| 23 – 30 | 1 (3.8%) |
| 30 – 59 | 10 (38.5%) |
| ≥ 60 | 15 (57.7%) |
| Sex | |
| Male | 13 (50%) |
| Female | 13 (50%) |
| Ward | |
| Internal ED | 22 (84.6%) |
| Surgical ED | 4 (15.4%) |
The relationship between the ability of biofilm formation and some risk factors including: underlying disease, previous hospitalization, antibioitics consumption and usage of different catheters, were evaluated and summarized in Table 2. No significant relationships were found between colonization with biofilm–forming MRSA and previous hospitalization, antibioitics consumption, usage of urine or vein catheter. Relationship between biofilm formation and antibiotic susceptibility pattern is shown in Figure 3. We found significant relationship (P value < 0.001) between strong biofilm formation and susceptibility to tetracycline.
Relationship Between the Ability of Biofilm Formation of MRSA Isolates From Nasal Carrier and Clinical Background
| Clinical Background | Biofilm Formation | |||||
|---|---|---|---|---|---|---|
| Strong | Medium + weak | P value | Odds ratio | CI 95% | ||
| Lower | Upper | |||||
| Underlying disease | 1 (3.8%) | 18 (69%) | 0.047 | 0.074 | 1.09 | 165.97 |
| Antibiotic consumption | 1 (3.8%) | 6 (23%) | ~ 1 | 0.889 | 0.07 | 10.30 |
| Usage of vein catheter | 3 (11.5%) | 18 (69%) | ~ 1 | 0.667 | 0.05 | 8.19 |
| Usage of urine catheter | 1 (3.8%) | 9 (35%) | ~ 1 | 0.481 | 0.43 | 5.40 |
| Previous hospitalization | 2 (7.7%) | 12 (46%) | ~ 1 | 0.833 | 0.09 | 7.02 |
Relationship Between Biofilm Formation of MRSA From Nasal Carrier and Antibiotic Susceptibility
5. Discussion
Biofilm forming bacteria cause a wide range of human infections. The resistant to antimicrobial agents among bacteria growing in biofilm are 500 to 5000 times higher than their planktonic counterparts (11). Microbial biofilm can be a serious health problem for patients need to use catheterization. Bacteria in biofilm are protected from antibiotics due to presence of large amount of exopolysaccharides, expression of biofilm specific resistance genes and having the suitable condition for growing slowly (5). MRSA is a pathogen causes various infections which usually show resistance to many of antibiotics. On the other hand, biofilm formation consider as a reservoir of pathogen that make them resistance to antibiotic agent and cause chronic infection, so increase the rate of MRSA carrier in the community, particularly MRSA with the ability of biofilm forming is a matter of concern (12-14).
Our results showed significant relationship between underlying disease and colonization by biofilm–forming MRSA, but no significant relationships were found between colonization with biofilm–forming MRSA and previous hospitalization, antibiotics consumption, usage of urine or vein catheter. Auxiliadora Molina and et al reported all the MRSA strains that isolated from blood culture were biofilm formers (12). In a study that was conducted in China, the prevalence of biofilm-forming MRSA was reported 66% (15). In South Africa 37.8% of strong biofilm producer were MRSA (16) which is higher than the rate of strong biofilm producer in this study. There are different ways for evaluating biofilm formation ability such as: microtitre plate, a tube test, radiolabelling, Congo red agar plate test and confocal laser scanning microscopy. The most popular of them is crystal violet microtitre plate assay (17).
In our study, the result of crystal violet microtitre plate assay, show that all of MRSA isolates have the ability of biofilm formation, and more than 60% of them could be able to produce slime layer on CRA. Nearly 40% of isolates that had recognized as a biofilm producer by crystal violet microtitre plate assay, reported as negative slime producer in CRA, these results shows that crystal violet microtitre plate assay is more sensitive than CRA assay. Among strong biofilm producer MRSA isolates, most resistance was seen against erythromycin and all of them were sensitive to tetracyclin and amikacin. We also found significant relationships between producing strong biofilm and being sensitive to tetracycline. Most of the strong biofilm formation-MRSA isolates showed high resistance to clindamycin, ciprofloxacin, SXT and gentamycin.
There are some studies that mentioned to some evidences about anti-biofilm activity of macrolides against Gram-negative organism, but there are contrary information about the effect of macrolides in biofilm-formation of Gram-positive organisms that our results are in agree with them (18). Nevertheless, in a study on MRSA strains, which isolated from cystic fibrosis patients, macrolides and gentamicin were reported as the active agents against biofilm formation (12).
Ciprofloxacin is reported as an effective antibiotic against biofilm–forming bacteria (11), but our results showed high resistance to ciprofloxacin. A study was conducted in Egypt, which was about the effect of ciprofloxacin on bacterial adherence and biofilm formation, the results of this study showed that ciprofloxacin decreased biofilm synthesis by ≥60% (5). Particularly, ciprofloxacin is one of the most currently prescribed classes of antibiotics in both the hospital and community (19), so it can be a reason for increasing resistance to ciprofloxacin. Tetracyclines are protein synthesis inhibitors, broad-spectrum, bacteriostatic antibiotics that target the 30S ribosome and prevent binding of tRNA and effective against both Gram-positive and Gram-negative bacteria. Studies on the effectiveness of tetracyclines against biofilm infections recommend that these compounds may work best as preventative actions (20). Tetracycline seem to be good candidates for further investigations in the treatment of MRSA biofilms. The threat of MRSA infections results from not only the occurrence of multidrug resistance but also the emergence of bacteria that form strong biofilms. In our study the rate of MRSA nasal carriage was high and 34.6% of MRSA isolates had the ability to form strong and medium biofilm. Since biofilm–forming capacity increase the resistance to common use antibiotics, isolating biofilm-formation MRSA from nasal carrier that can be easily transmitted to other people in the community and hospital, is an alarming for public health.
References
-
1.
Ando E, Monden K, Mitsuhata R, Kariyama R, Kumon H. Biofilm formation among methicillin-resistant Staphylococcus aureus isolates from patients with urinary tract infection. Acta Med Okayama. 2004;58(4):207-14. [PubMed ID: 15551758].
-
2.
Gowrishankar S, Duncun Mosioma N, Karutha Pandian S. Coral-Associated Bacteria as a Promising Antibiofilm Agent against Methicillin-Resistant and -Susceptible Staphylococcus aureus Biofilms. Evid Based Complement Alternat Med. 2012;2012:862374. [PubMed ID: 22988476]. https://doi.org/10.1155/2012/862374.
-
3.
Atshan SS, Shamsudin MN, Lung LT, Sekawi Z, Ghaznavi-Rad E, Pei CP. Comparative characterisation of genotypically different clones of MRSA in the production of biofilms. J Biomed Biotechnol. 2012;2012:417247. [PubMed ID: 22529705]. https://doi.org/10.1155/2012/417247.
-
4.
Del Papa MF, Hancock LE, Thomas VC, Perego M. Full activation of Enterococcus faecalis gelatinase by a C-terminal proteolytic cleavage. J Bacteriol. 2007;189(24):8835-43. [PubMed ID: 17921295]. https://doi.org/10.1128/JB.01311-07.
-
5.
El-Feky MA, El-Rehewy MS, Hassan MA, Abolella HA, Abd El-Baky RM, Gad GF. Effect of ciprofloxacin and N-acetylcysteine on bacterial adherence and biofilm formation on ureteral stent surfaces. Pol J Microbiol. 2009;58(3):261-7. [PubMed ID: 19899620].
-
6.
Murugan K, Usha M, Malathi P, Al-Sohaibani AS, Chandrasekaran M. Biofilm forming multi drug resistant Staphylococcus spp. among patients with conjunctivitis. Pol J Microbiol. 2010;59(4):233-9. [PubMed ID: 21466040].
-
7.
Mataraci E, Dosler S. In vitro activities of antibiotics and antimicrobial cationic peptides alone and in combination against methicillin-resistant Staphylococcus aureus biofilms. Antimicrob Agents Chemother. 2012;56(12):6366-71. [PubMed ID: 23070152]. https://doi.org/10.1128/AAC.01180-12.
-
8.
Performance Standards for Antimicrobial Susceptibility Testing. Twenty-First Informational Supplement. 2011;31(1).
-
9.
Ghaznavi-Rad E, Nor Shamsudin M, Sekawi Z, van Belkum A, Neela V. A simplified multiplex PCR assay for fast and easy discrimination of globally distributed staphylococcal cassette chromosome mec types in meticillin-resistant Staphylococcus aureus. J Med Microbiol. 2010;59(Pt 10):1135-9. [PubMed ID: 20616192]. https://doi.org/10.1099/jmm.0.021956-0.
-
10.
Seno Y, Kariyama R, Mitsuhata R, Monden K, Kumon H. Clinical implications of biofilm formation by Enterococcus faecalis in the urinary tract. Acta Med Okayama. 2005;59(3):79-87. [PubMed ID: 16049560].
-
11.
El-Shekh NA, Ayoub AMA, El-Hendawy HH, Abada EA, Khalifa SYE. In vitro Activity of some Antimicrobial Agents against Intact and Disrupted Biofilms of Staphylococci in the Indwelling Vascular Catheter Patients. World Appl Sci J. 2010;10(1):108-120.
-
12.
Molina A, Del Campo R, Maiz L, Morosini MI, Lamas A, Baquero F, et al. High prevalence in cystic fibrosis patients of multiresistant hospital-acquired methicillin-resistant Staphylococcus aureus ST228-SCCmecI capable of biofilm formation. J Antimicrob Chemother. 2008;62(5):961-7. [PubMed ID: 18647744]. https://doi.org/10.1093/jac/dkn302.
-
13.
Sasirekha B, Usha MS, Amruta AJ, Ankit S. Evaluation and Comparison of Different Phenotypic Tests to Detect Methicillin Resistant Staphylococcus aureus and their Biofilm Production. Int J PharmTech Res. 2012;4(2):532-541.
-
14.
O'Neill E, Pozzi C, Houston P, Smyth D, Humphreys H, Robinson DA, et al. Association between methicillin susceptibility and biofilm regulation in Staphylococcus aureus isolates from device-related infections. J Clin Microbiol. 2007;45(5):1379-88. [PubMed ID: 17329452]. https://doi.org/10.1128/JCM.02280-06.
-
15.
Wang L, Yu Fangyou, Yang Lehe, Li Qiaoqiao, Zhang Xueqing, Zeng Yunxiang, et al. Prevalence of virulence genes and biofilm formation among Staphylococcus aureus clinical isolates associated with lower respiratory infection. Afr J Microbiol Res. 2010;4:2566-2569.
-
16.
Samie A, Shivambu N. Biofilm production and antibiotic susceptibility profiles of Staphylococcus aureus isolated from HIV and AIDS patients in the Limpopo Province, South Africa. Afr J Biotechnol. 2011;10(65):14625-14636.
-
17.
Kawamura H, Nishi J, Imuta N, Tokuda K, Miyanohara H, Hashiguchi T, et al. Quantitative analysis of biofilm formation of methicillin-resistant Staphylococcus aureus (MRSA) strains from patients with orthopaedic device-related infections. FEMS Immunol Med Microbiol. 2011;63(1):10-5. [PubMed ID: 21595755]. https://doi.org/10.1111/j.1574-695X.2011.00821.x.
-
18.
Parra-Ruiz J, Vidaillac C, Rybak MJ. Macrolides and staphylococcal biofilms. Rev Esp Quimioter. 2012;25(1):10-6. [PubMed ID: 22488536].
-
19.
Petrelli D, Repetto A, D'Ercole S, Rombini S, Ripa S, Prenna M, et al. Analysis of meticillin-susceptible and meticillin-resistant biofilm-forming Staphylococcus aureus from catheter infections isolated in a large Italian hospital. J Med Microbiol. 2008;57(Pt 3):364-72. [PubMed ID: 18287301]. https://doi.org/10.1099/jmm.0.47621-0.
-
20.
Kiedrowski MR, Horswill AR. New approaches for treating staphylococcal biofilm infections. Ann N Y Acad Sci. 2011;1241:104-21. [PubMed ID: 22191529]. https://doi.org/10.1111/j.1749-6632.2011.06281.x.