Abstract
Background:
Lipases (acylglycerol acylhydrolase, E. C. 3. 1. 1. 3) are widely distributed among microorganisms, animals and plants, catalyzing the hydrolysis of triglycerides to free fatty acids and glycerol. Their commercial application includes pharmaceutical, chemical, and paper industries.Objectives:
This study aimed to isolate and screen lipolytic fungi from coastal waters of the southern Caspian Sea by Internal Transcribed Spacer-Polymerase Chain Reaction (ITS-PCR), and to optimize their lipolytic activity, pH and temperature. The ITS regions possess a high variation among taxonomically distinct fungal species and even within species.Materials and Methods:
All fungal were tested to determine their lipolytic activity by the Tributyrin agar plate assay. After DNA extraction, lipase-producing fungi were identified via ITS-PCR of rDNA region with ITS1 and ITS4 primers.Results:
Four fungal species were isolated from water samples of the Caspian Sea (north of Iran) between February and June 2011. The nucleotide sequences reported for three of these isolates have been assigned accession numbers from NCBI Gene Bank database. Among these species, Cladosporium langeronii showed maximum lipolytic activity (34 U/mL) and maximum clear zone formation (6 mm) on the Tributyrin agar plates. The optimum pH and temperature for activity were 8.0 and 35°C, respectively.Conclusions:
The findings of this study indicated that these isolates were plant pathogenic fungi, which entered seawater from the environment, and were likely to have a suitable lipase activity on plant oils.Keywords
1. Background
Caspian Sea is amongst valuable water resources, which takes up nearly 40% of the total area of lakes in the world and because of the lack of relationships with other oceans, it has become a unique aquatic habitat. The physicochemical conditions of this lake, especially its salt concentration, have an important role in creating its unique biodiversity (1). Among organisms in aquatic ecosystems, fungi have efficient enzymatic systems, and have important effects on biodegradation of undesirable compounds, including industrial and agricultural effluents, detergents and various oils (2, 3). Although fungi have been identified in aquatic environments for many years, their importance is often ignored in the development of ecological processes and the elimination of water pollution (3).
Lipases (triacylglycerol acylhydrolase, EC 3. 1. 1. 3) are hydrolytic enzymes that catalyze the hydrolysis of fats and oils to glycerol and free fatty acids at the oil-water interface (4). Recently, lipases have been the subject of active research on account of their versatile applications in different industries. Lipases with new properties are of particular interest, and can be obtained through isolation from various natural sources (5). Although lipases have been widely used in several areas of medicinal, biotechnological, detergent, dairy, and food and hydrolysis of fats and oil, there are still substantial current interests in developing new microbial lipases (6, 7). A considerable number of fungal and bacterial lipases have been commercially produced with the former being preferable, because fungi generally produce extracellular enzymes which facilitate recovery of the lipase from the fermentation broth (8).
Internal Transcribed Spacer (ITS) regions have been successfully used to generate specific primers capable of differentiating closely related fungal species. Analysis of the ITS is also sensitive enough to identify intraspecific variation that is not apparent from morphological analysis (9). In the present paper, we did not only determine lipase activity at different temperatures and pH values, but also performed molecular identification of these species by the ITS-PCR sequencing method.
2. Objectives
The Caspian Sea has a high diversity of marine fungi and different species of fungi are able to produce valuable enzymes. The objective of the present study was to isolate and screen lipolytic fungi from coastal waters of the southern Caspian Sea by ITS-PCR, and to optimize their lipolytic activity, pH and temperature.
3. Materials and Methods
3.1. Isolation of Microorganisms
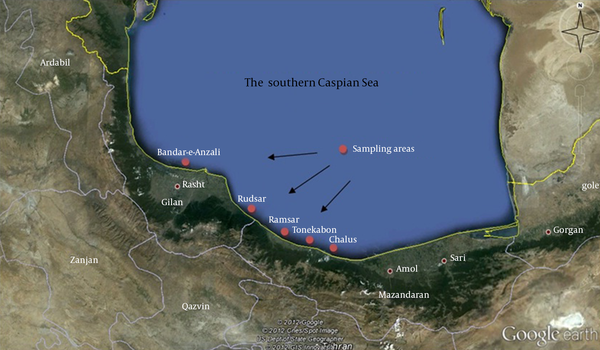
Gilan and Mazandaran provinces in north of Iran are located on the southern coasts of the Caspian Sea. This region possesses a moderate climate during all seasons with plenty of rainfall. In this study, sampling was conducted from Chalus, Tonekabon, Ramsar, Rudsar and Bandar-e-Anzali cities (Figure 1). Between February and June 2011, water samples were collected in 50 mL sterile glass bottles, from different regions of the southern Caspian Sea from sections, which had stagnant waters. Next, the water samples were transported to the laboratory at ambient temperature and stored at 4°C before they were analyzed within two hours. Serial dilutions were prepared and 100 μL from each dilution was cultivated on Potato Dextrose Agar (PDA) (Merck, Germany) and incubated at 30°C for five days. Grown fungi colonies were purified on PDA plates (10).
The Coastal Areas of Water Sample Collection
3.2. Microscopic Identification of Fungi
For initial detection of fungal samples direct observation of samples was used. In microscopic examination, slides were observed after staining samples with Lactophenol Cotton Blue (3).
3.3. Detection of Lipase Activity by Tributyrin Agar Plate Assay
We used the sensitive plate assay for detection of lipase activity in growing cultures (10, 11). Detection of lipase activity was performed on Tributyrin agar (Merck, Germany) containing glycerol oil as the substrate to identify H/C for each isolate. The H/C clearly illustrates the ratio of the diameters of hydrolysis halo on Tributyrin agar (H) relative to cell colony size (C). An H/C ratio of one means no hydrolysis, thus no lipase production, whereas a higher H/C ratio indicates lipase secretion. Isolated fungi were cultured on Tributyrin plate.and incubated at 30°C for 48 hours. The colonies showing clear zones were picked from plates and inoculated in Tributyrin broth for quantitative determination of lipase activity. The pH of the medium was adjusted to 7.0, using 0.1 M NaOH and incubated in a rotary shaker (100 rpm) at 30°C for 96 hours.
3.4. DNA Extraction
The strains which showed maximum lipolytic activity were identified by internal transcribed spacer (ITS) sequencing. The genomic DNA was isolated according to the procedure described by Lian et al. (12). Briefly, 400 μL of lysis buffer (100 mM NaCl, 25 mM EDTA, 0.5% sodium dodecyl sulfate (SDS), and 200 mM Tris-HCl) was mixed with 50 mg of fungal tissue. After incubation at 65°C for one hour, 350 μL of Cetyltrimethyl Ammonium Bromide (CTAB) buffer (5% CTAB [w/v], 100 mM Tris-HCl pH 8.0, 1.4 M NaCl) was added and incubated for 10 minutes in a water bath. Then, 700 μL of the mixture of phenol/chloroform/isoamyl alcohol (1: 24: 25) was added to the test tube, followed by centrifugation at 12,000 rpm. Then the same amount of the supernatant chloroform/amyl alcohol was added and centrifuged. After transition of supernatant to a new tube, an equal volume of isopropanol was added and centrifuged at maximum rpm. The tube was washed with cold 70% ethanol and the pellet of DNA was air-dried for five minutes and re-suspended in 40 µL of sterile water. The DNA was either used immediately or frozen at -20˚C for later analysis.
3.5. Polymerase Chain Reaction Amplification and Sequencing of Partial Sequence of Lipase Gene
The rDNA gene was amplified, using the two primers ITS1 (5´-TCC GTA GGT GAA CCT GCG G-3´) and ITS4 (5´-TCC TCC GCT TAT TGA TAT GC-3´) (Gene Fanavaran., Iran) based on the protocol described by Ferrer et al. (13) with some minor modifications. A total of 3 µL of purified DNA was used as the template for PCR (BIO-RAD, Germany). The PCR reaction volume was 50 μL containing 25 pmol of each primer, 3 mM MgCl2, 200 µM dNTPs, and 2.5 U Taq DNA polymerase (Cinagen, Tehran, Iran) in 10x reaction buffer. The PCR cycling conditions included 95˚C for five minutes (1 cycle), 95˚C for 30 seconds, 55˚C for one minute, 72˚C for one minute (35 cycle), and 72˚C for six minutes (1 cycle). After amplification, a volume of 10 µL of the PCR product was run on 1% agarose gel by electrophoresis and bands were visualized using a UV trans-illuminator (Uvitec, United Kingdom). A negative control (sterile water) was used in each of the reactions. Finally, PCR amplified fragments were sequenced (Macrogen, Korea). The deduced sequence was subjected to the Basic Alignment Search Tool (BLAST) for the closet match in the database (14).
3.6. Optimization Experiments
Lipase was produced by growing the fungal species in Tributyrin broth medium containing yeast extract (3 g/L); peptone (5 g/L) and 10% (v/v) glycerol. The same medium was used to optimize lipase production. Lipase production was optimized by varying pH (4-9) and temperature (25 - 65°C).
3.7. Enzyme Activity Assay
Lipase activity was determined by a spectrometric method using p-nitrophenyl palmitate (p-NPP) (Sigma, USA) as the substrate with some minor modifications (7, 15). Activity measurements were carried out by determining the absorbance of liberated p-nitrophenyl at 410 nm, based on the protocol of Maia et al. (15). The assay mixture consisted of 200 µL of sample and 800 µL of substrate solution containing 3 mg of pNPP dissolved in 1 mL of 2-propanol diluted in 9 mL of 50 mM Tris-HCl pH 8.0 with 20 mg of Triton X-100 mg and 1 mg of glycerol. The assay mixture was incubated at 40˚C for one hour. One unit of activity was defined as the amount of enzyme that liberated 1 nmol of p-nitrophenyl per minute under the assay conditions. This method was the standard assay for lipase activity.
3.8. Effects of pH on Lipase Activity
The optimum pH for the purified lipase was determined by incubating the enzyme-substrate at various pH values ranging from 4 to 9 at room temperature in the following buffers: citrate-phosphate pH 6.0 and 7.0; phosphate pH 7.0 and 8.0; Tris-HCl pH 8.0 and 9.0 and glycine-NaOH pH 9.0 and 10.0, all at 50 mM (8).
3.9. Effect of Temperature on Lipase Activity
The optimum temperature for the purified lipase was determined by incubating the reaction mixture at temperatures ranging from 25˚C to 65˚C at the optimized pH value.
4. Results
4.1. Isolation and Identification of Lipolytic Fungi
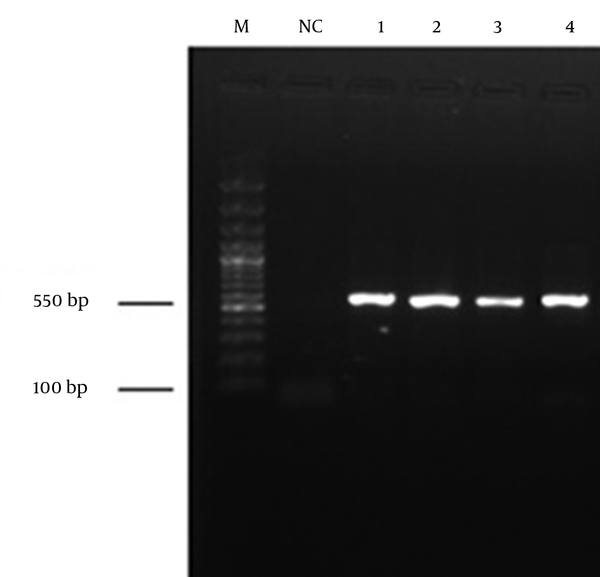
From the total samples obtained from coastal waters of the southern Caspian Sea (Chalus, Tonekabon, Ramsar, Rudsar and Bandar-e-Anzali in the north of Iran), four fungal species were isolated and identified. The total size of the ITS1 and ITS2 regions, including the 5.8S rDNA gene, of the isolates was 550 base pairs (bp). The ITS sequencing data indicated that the isolates were Cladosporium langeronii, Phaeosphaeria setosa, C. cladosporioides and Glomerella acutata (Figure 2). The nucleotide sequences reported here for C. langeronii, Phaeosphaeria setosa and C. cladosporioides have been assigned accession numbers of JF810856, JF810857 and JF810858, respectively by the NCBI Gene Bank database. The BLAST results showed 98% similarity for C. langeronii and 99% for P. setosa and C. cladosporioides with other fungal species (Figures 3, 4 and 5). Among the four isolated species, only C. langeronii showed maximum lipolytic activity (34 U/mL) and maximum clear zone formation (6 mm) on the Tributyrin agar plates containing glycerol substrate, indicating the production of an extracellular lipase, and for this reason C. langeronii was selected for the investigation of pH and temperature.
Polymerase Chain Reaction Products Obtained From Water Samples Following Internal Transcribed Spacer-Polymerase Chain Reaction. Lanes 1-4, PCR products of fungal species with a specific amplification product of 550 bp; NC, negative control; M, molecular weight ladder (100 - 3000 bp).
Blast Results of Cladosporium langeronii With an Accession Number of JF810856 From NCBI Gene Bank

Blast Results of Phaeosphaeria setosa With an Accession Number of JF810857 From NCBI Gene Bank

Blast Results of Cladosporium Cladosporioides With an Accession Number of JF810858 From NCBI Gene Bank

4.2. Effect of pH and Temperature on Lipase Production
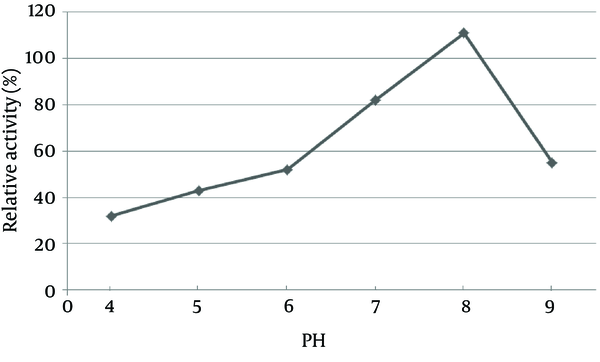
The pH of the culture is one of the most important environmental parameters that affect microbial cell growth and enzyme production. The effect of pH on lipase activity of C. langeronii was determined for pH values ranging from four to nine as shown in Figure 6. Maximum lipase activity of C. langeronii was obtained when the initial pH of the growth medium was 8.0, in the Tris-HCl buffer. The incubation temperature of the growth media is also one of the critical factors for lipase production, where the optimum temperature varies from one organism to other. The optimal temperature for lipase activity by C. langeronii in the present study was 35˚C and high activity was obtained between 35˚C and 45˚C (Figure 7). The sharp decrease in lipase activity at higher temperatures may be due to the denaturation effect of the produced lipase.
Effect of pH on Lipase Production by Cladosporium langeronii After 48 Hours at 35°C
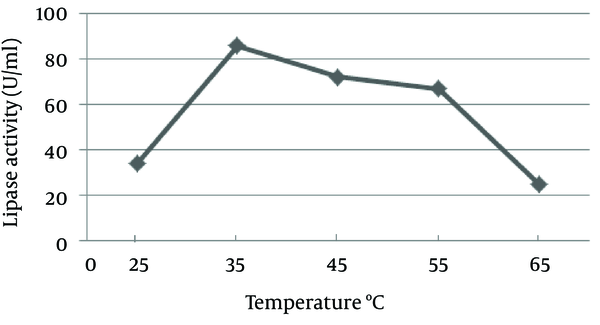
Effect of Temperature on Lipase production by Cladosporium langeronii After 48 Hours (pH 8.0)
5. Discussion
Fungi cause most soil-borne vegetable diseases and thus are considered the most important pathogenic group of organisms. Plant-pathogenic fungi fall into five main taxonomic classes based on morphological and biological characteristics: Plasmodiophoromycetes, Zygomycetes, Oomycetes, Basidiomycetes and Ascomycetes (16). Our results indicated that most strains isolated from coastal waters of the southern Caspian Sea, belonged to the genus of Cladosporium and the series of Ascomycota. Also, Damare et al. (17) showed that genuses of Penicillium, Aspergillus and Cladosporium are the most abundant fungal species in aquatic environments. Furthermore, spores of Cladosporium have been often seen in aquatic environments, which are located in the vicinity of agricultural land and farms (18).
Since the main habitat of this fungus is the soil, it is able to adapt, grow and survive in aquatic environments, with production of extracellular enzymes (2, 3). In a closed ecosystem like the Caspian Sea, with the salt concentration and lack of nutrients, marine fungi could be a good source for industrial applications Also in the meantime, we should consider oily wastewater pollution that enters the lake. Vegetable oils that are used for different applications are released and accumulate in the environment. A few studies that have been done on oily wastewater, indicated that different fungal species such as Cladosporium with production of extracellular enzymes such as lipases, are very useful in decomposition of oil and cleaning up the environment (2). The identification of fungal species by classic taxonomy is based mainly on the use of morphological markers. However, the number of available markers is generally low, which makes the classification or identification of related species difficult.
The amplification of Internal Transcribed Spacer (ITS) of ribosomal DNA (rDNA) by the Polymerase Chain Reaction (PCR), combined with sequencing of the amplicon and analysis of similarity between the obtained sequences and those already deposited in the gene bank data base, has been frequently employed for identification of fungal species. The rRNA gene for 5.8S RNA separates the two ITS regions. The sequence variation of ITS regions has led to their use in phylogenetic studies of many different organisms. Recently, Guarro et al. (19) proposed the use of ITS amplicons of different lengths for identification of Aspergillus species by capillary electrophoresis. The focal theme of the present investigation was to produce lipase from fungi, which were isolated from coastal waters of the southern Caspian Sea. These species were identified using ITS-PCR sequencing and microscopic analysis.
Our experiments on the effect of temperature on activity of lipase were confined to the range of 20 - 65°C. The optimal temperature of 35°C was similar to that of lipase from Cryptococcus sp. S-2, described by Kamini et al. (20). Like most fungal and yeast lipases that generally have optimal activity at neutral pH, the lipase from C. langeronii had maximum activity at pH 8.0 and 35˚C (8). High pH optima for lipase activity have also been reported for Antrodia cinnamomea and Rhizopus oryzae (21, 22). Similar results were reported by Gopinath et al. (23) and Yu et al. (24) for lipase from Cunninghamellaverticillata and Yarrowia lipolytica with maximum activities at pH 7.5 and 8.0, respectively. Since the lipase from C. langeronii retained 51% and 57% activity even at pH 6.0 and 9.0, respectively, it had good alkaline lipase characteristics for potential application in decomposition of oil and cleaning up the environment.
Acknowledgements
References
-
1.
Zeinolabedin Y, Yahyapour MS, Shirzad Z. Geopolitics and Environmental Issues in the Caspian Sea. Caspian J Environ Sci. 2009;7(2):113-21.
-
2.
Gopinath SCB, Hilda A, Anbu P. Extracellular enzymatic activity profiles in fungi isolated from oil-rich environments. Mycoscience. 2005;46(2):119-26.
-
3.
Das S, Lyla PS, Khan SA. Filamentous fungal population and species diversity from the continental slope of Bay of Bengal, India. Acta Oecol. 2009;35(2):269-79. https://doi.org/10.1016/j.actao.2008.11.003.
-
4.
Liu R, Jiang X, Mou H, Guan H, Hwang H, Li X. A novel low-temperature resistant alkaline lipase from a soda lake fungus strain Fusarium solani N4-2 for detergent formulation. Biochem Eng J. 2009;46(3):265-70. https://doi.org/10.1016/j.bej.2009.05.016.
-
5.
Svendsen A. Lipase protein engineering. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 2000;1543(2):223-38.
-
6.
Lin ES, Wang CC, Sung SC. Cultivating conditions influence lipase production by the edible Basidiomycete Antrodia cinnamomea in submerged culture. Enzyme Microb Technol. 2006;39(1):98-102. https://doi.org/10.1016/j.enzmictec.2005.10.002.
-
7.
Supakdamrongkul P, Bhumiratana A, Wiwat C. Characterization of an extracellular lipase from the biocontrol fungus, Nomuraea rileyi MJ, and its toxicity toward Spodoptera litura. J Invertebr Pathol. 2010;105(3):228-35. [PubMed ID: 20600093]. https://doi.org/10.1016/j.jip.2010.06.011.
-
8.
Lima VMG, Krieger N, Mitchell DA, Fontana JD. Activity and stability of a crude lipase from Penicillium aurantiogriseum in aqueous media and organic solvents. Biochem Eng J. 2004;18(1):65-71.
-
9.
Abd-Elsalam KA, Aly IN, Abdel-Satar MA, Khalil MS, Verreet JA. PCR identification of Fusarium genus based on nuclear ribosomal-DNA sequence data. Afr J Biotechnol. 2004;2(4):82-5.
-
10.
Cardenas F, de Castro MS, Sanchez-Montero JM, Sinisterra JV, Valmaseda M, Elson SW, et al. Novel microbial lipases: catalytic activity in reactions in organic media. Enzyme Microb Technol. 2001;28(2-3):145-54. [PubMed ID: 11166805].
-
11.
Mafakher L, Mirbagheri M, Darvishi F, Nahvi I, Zarkesh-Esfahani H, Emtiazi G. Isolation of lipase and citric acid producing yeasts from agro-industrial wastewater. N Biotechnol. 2010;27(4):337-40. [PubMed ID: 20450991]. https://doi.org/10.1016/j.nbt.2010.04.006.
-
12.
Lian B, Zang JP, Hou WG, Yuan S, Smith DL. PCR-based sensitive detection of the edible fungus Boletus edulis from rDNA ITS sequences. Electron J Biotechnol. 2008;11(3):102-9.
-
13.
Ferrer C, Colom F, Frases S, Mulet E, Abad JL, Alio JL. Detection and identification of fungal pathogens by PCR and by ITS2 and 5.8S ribosomal DNA typing in ocular infections. J Clin Microbiol. 2001;39(8):2873-9. [PubMed ID: 11474006]. https://doi.org/10.1128/JCM.39.8.2873-2879.2001.
-
14.
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2006;34(Database issue):D16-20. [PubMed ID: 16381837]. https://doi.org/10.1093/nar/gkj157.
-
15.
Maia MM, Heasley A, Camargo de Morais MM, Melo EH, Morais MJ, Ledingham WM, et al. Effect of culture conditions on lipase production by Fusarium solani in batch fermentation. Bioresour Technol. 2001;76(1):23-7. [PubMed ID: 11315806].
-
16.
Shearer CA, Descals E, Kohlmeyer B, Kohlmeyer J, Marvanova L, Padgett D, et al. Fungal biodiversity in aquatic habitats. Biodivers Conserv. 2007;16(1):49-67.
-
17.
Damare S, Raghukumar C, Raghukumar S. Fungi in deep-sea sediments of the Central Indian Basin. Deep Sea Res Part 1 Oceanogr Res Pap. 2006;53(1):14-27. https://doi.org/10.1016/j.dsr.2005.09.005.
-
18.
Panda T, Panda B, Mishra N. Seasonal incidence of air borne fungi in coastal belt of Orissa. J Hum Ecol. 2009;26(3):205-7.
-
19.
Guarro J, Gené J, Stchigel AM. Developments in fungal taxonomy. Clin Microbiol Rev. 1999;12(3):454-500.
-
20.
Kamini NR, Fujii T, Kurosu T, Iefuji H. Production, purification and characterization of an extracellular lipase from the yeast, Cryptococcus sp. S-2. Process Biochem. 2000;36(4):317-24.
-
21.
Shu CH, Xu CJ, Lin GC. Purification and partial characterization of a lipase from Antrodia cinnamomea. Process Biochemistry. 2006;41(3):734-8. https://doi.org/10.1016/j.procbio.2005.09.007.
-
22.
Minning S, Schmidt-Dannert C, Schmid RD. Functional expression of Rhizopus oryzae lipase in Pichia pastoris: high-level production and some properties. J Biotechnol. 1998;66(2-3):147-56. [PubMed ID: 9866866].
-
23.
Gopinath SCB, Hilda A, priya TL, Annadurai G. World J Microbiol Biotechnol. 2002;18(5):449-58.
-
24.
Yu M, Qin S, Tan T. Purification and characterization of the extracellular lipase Lip2 from Yarrowia lipolytica. Process Biochem. 2007;42(3):384-91. https://doi.org/10.1016/j.procbio.2006.09.019.