Abstract
Background:
Long-term lamivudine therapy, despite its initial effectiveness against hepatitis B virus (HBV), is associated with the emergence of drug resistance mutations in polymerase protein.Objectives:
The aim of the present study was to determine the prevalence of precore and lamivudine drug resistance mutations in lamivudine treated patients with chronic B hepatitis.Patients and Methods:
Sequential sera were obtained from 88 chronic HBV carriers who received lamivudine for more than 24 months. Polymerase and precore regions were directly sequenced for these groups: I (before treatment), II, and III (12 and 24 months after treatment, respectively).Results:
All patients (100%) were contained genotype D, subtype ayw2. One (1.1%), 12 (13.6%), and 22 (25%) members of groups I, II, and III had the replacement of either isoleucine or valine instead of methionine in tyrosine-methionine-aspartate-aspartate (YMDD) motif, respectively. The frequency of mutations from 0 time point to 12 and 24 months showed that there was an increasing trend between sequential samples (P < 0.001). In group I, 31 (35.2%); II, 36 (41.0%) and III, 41 (46.6%) members had the precore stop codon mutations. The frequency of mutations from 0 time point to 12 and 24 months showed that there was an ascending trend between sequential samples. Indeed, frequency of precore stop codon was significantly increased with the passage of time (P < 0.001).Conclusions:
Presence of drug resistance mutations among the patients was significant. Precore mutations were common amongst Iranian HBV chronic carriers under lamivudine therapy and these mutations were accompanied by clinical relapse.Keywords
1. Background
Hepatitis B virus is the leading cause of liver disease, including fulminant hepatitis, cirrhosis, and hepatocellular carcinoma (HCC). The latter complication is at the top of ten leading causes of death worldwide. Globally, more than 400 million chronic carriers are at the risk for such complication (1). Currently treatment options rely on Interferon alpha (INF-α) and Nucleos(t)ide Analogues (NA) (2). Lamivudine (LAM) was the first approved oral NA, which was effective on HBV replication. It entered into the market in 1998 for widespread clinical use. It can remarkably reduce serum HBV DNA levels, enhance hepatitis B e antigen (HBeAg) seroconversion, thus decreasing the progression of liver disease (3, 4). LAM is a synthetic NA that unlike IFN, does not have immunomodulatory effects, however, there is evidence to suggest that lamivudine treatment may overcome cytotoxic T cell hyporesponsiveness seen in chronically infected patients (5).
In spite of this, prolonged LAM treatment is associated with emergence of the classic lamivudine-resistance in the tyrosine-methionine-aspartate-aspartate (YMDD) motif of HBV polymerase, which is the substitution of either valine or isoleucine for methionine (rtM204I/V, domain C) and upstream compensatory mutations in domains A(L80V/I, L82M, L91I) and B (rtV173L, rtL180M) of the reverse transcriptase (3, 6). Genotypic resistant mutations have been detected in 14% - 32% of patients after one year of treatment, reaching 70% after 4 years of treatment for lamivudine (7, 8). Emergence of the lamivudine-resistant variants may be accompanied with virological and biochemical relapses followed by acute exacerbation of liver disease and although rare, hepatic decompensation (9). Although LAM is more convenient than interferon-based therapies along with fewer side effects, prolonged viral suppression is usually not achieved after a 48-week course of therapy, which require long, and in many cases, indefinite treatment. Alas, extended LAM treatment is associated with an elevated risk of developing drug resistance. The reported prevalence of YMDD variants in Iranian patients undergone LAM therapy ranged between 14% and 33% (10-12).
In populations worldwide, variants of HBV are selected after seroconversion to anti-HBe a translational stop codon at the 3’ end of precore, predicting the prevention of HBeAg synthesis, was reported in several studies on chronic HBV-infected patients (13). Precore variant has been described in asymptomatic carriers (14), fulminant hepatitis (15, 16), patients with hepatocellular carcinoma (17, 18), after interferon (IFN) therapy (19, 20), and following immunization (21). This variant appears to be associated with severe liver disease in HBeAg negative patients with enhanced HBV replication (22). Precore mutant has been reported at a range of 38% to 58% among Iranian chronic carriers (23-25).
2. Objectives
The aims of the present study were (i) to determine the prevalence of YMDD and precore mutations from sequential samples obtained from CHB patients undergone lamivudine therapy, and (ii) to explore the relationship between virological breakthrough and mutational patterns in these patients.
3. Patients and Methods
A total of 88 HBsAg-positive chronic carrier patients referred to the digestive disease research center in Shariati hospital, Tehran were enrolled in a cross-sectional retrospective study. All patients were interviewed and examined by gastroenterologists to evaluate the clinical findings and the results of the investigative workup (liver histology, ultrasonography, and laboratory tests such as serologic, biochemical, and virological tests) in order to determine the clinical status of the patient. Chronic hepatitis was defined as HBsAg positivity with or without the presence of HBeAg and moderate to high HBV DNA levels, persistent or intermittent elevation in the serum ALT levels, and compatible liver biopsy.
The selected patients did not have HDV, HIV, and HCV co-infection or receive any medication or vaccine. All patients were given their informed consent and the study protocol was approved by the local ethics committee (No. 3954). The eligible patients received 100 mg LAM orally. The patients were divided into three groups: I: before treatment, II, and III, 12 and 24 months after initiation of treatment, respectively. Five milliliter aliquots of whole blood samples were withdrawn from each participant. Serum was aseptically separated in the field by centrifugation at 2000 rpm for 5 min and stored at -20°C until be tested. HBV serological markers, including HBsAg and anti-HBs were examined by ELISA kits manufactured by Organon (Technika, The Netherlands).
3.1. DNA Extraction
HBV DNA was extracted from a 200 µL of sera using Qiagen mini blood kit (QIAGEN, Hilden, Germany) according to manufacturer’s instruction. In brief, 20 µL of protease added to the serum in a 1.5 mL tube. Then, 200 µL of Al buffer added to each tube, were vortexed and incubated for 10 min at 56°C. For DNA precipitation, 200 µL of ethanol was added to the mixture, centrifuged for 1 min. Components were transferred to a collection tube contained filter. Trapped DNA was washed in two steps by AW1 and AW2 buffers to eliminate impurities together with centrifugation after each step. Finally, DNA was eluted using 100 µL of elution buffer, stored at -20°C.
3.2. Polymerase Chain Reaction
Specific primers were used for the amplification of RT (26) and precore (27) regions, as described previously. For the purpose of HBV genotyping, nested PCR on both genes was carried out as mentioned elsewhere (28). Five microliter of extracted HBV DNA was used for the first round and 1 µL of the first round amplicon was utilized for the second round PCR reaction as the template.
3.3. DNA Sequencing
The sequencing analysis of the whole surface antigen region obtained from the second stage PCR was done with a DNA sequence analyzer (Perkin Elmer ABI-3130XL DNA Sequencer, Foster City, CA, USA) using 0.5 μL of appropriate internal primers as described previously (26, 27). Electropherograms were analyzed with the Chromas program and checked manually to confirm base assignment.
3.4. Sequence Analysis
For the purpose of sequencing alignment, after allocating a sequence to an HBV genotype by analysis of the S gene, the found variations of surface gene amino acid/nucleotide were compared with reference sequences obtained from available international sequences and the consensus HBsAg sequences from Iranian isolates obtained from GenBank, NCBI and from our laboratory using BioEdit package version 7.0.9. Comparing to the former, any amino acid changes defined as “variant” (host HLA-determined). With regard to the latter (Iranian database sequences), amino acid differences defined as “mutation”.
3.5. Statistical Analysis
Data were analyzed with SPSS 16.0 (SPSS Inc., Chicago, Illinois, USA). Baseline descriptive data were expressed as mean ± SD for continuous variables and as frequencies and percentages for categorical variables. Associations were analyzed with Student t-test and categorical variables by Chi-square or Fisher exact test. For all comparisons, statistical significance was determined at 0.05 level.
4. Results
4.1. Demographic and Clinical Characteristics
The baseline characteristics and sequential changes of mutational patterns before and after therapy (12 and 24 months later) are shown in Table 1. There were 64 (72.7%) males and 24 (27.3%) females, and the mean age of the patients was 38.02 years (range, 16 - 71). At the time of presentation, all participants were HBeAg negative and anti-HBe positive. From time point 0 to 24 (months), the levels of HBV DNA and ALT/AST in patients showed a significant decline (P values < 0.001 and < 0.002, respectively).
| Variable | Group I | Group II | Group III | P Value |
|---|---|---|---|---|
| Age, y | 38 (16-71) | 39 (17-72) | 40 (18-73) | > 0.05 |
| Gender | ||||
| Male | 64 (72.7) | |||
| Female | 24 (27.3) | |||
| HBeAg negative | 88 (100) | 88 (100) | 88 (100) | - |
| Anti-HBe positive | 88 (100) | 86 (97.7) | 86 (97.7) | - |
| ALT, IU/L | 115 ± 72.5 | 37.6 ± 22.2 | 42.3 ± 45 | < 0.001 |
| AST, IU/L | 70 ± 54.5 | 31. ± 15.5 | 35.5 ± 36 | 0.002 |
| HBV-DNA, Log10 IU/mL | 5.037 ± 1.5 | 1.41 ± 1.6 | 1.84 ± 2.3 | < 0.001 |
4.2. Mutational Analysis
Analysis of variation within the S gene of the patients with chronic HBV infection demonstrated that the only detected genotype was D (100%) and subtype ayw2 (100%).
4.3. RT Domain
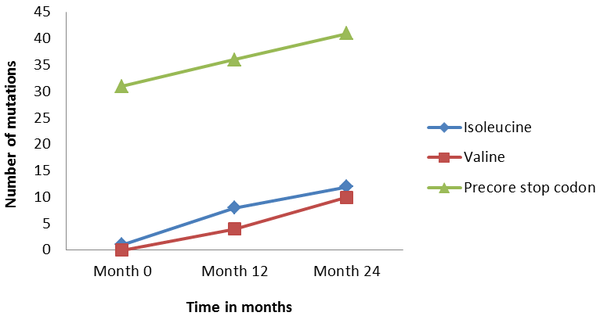
Tables 1 and 2 present the alignment of deduced amino acid sequences of the RT domain of polymerase protein obtained from the serial serum samples before administration of LAM and after treatment. In group I, one (1.1%); in group II, 12 (13.6%), and in group III, 22 (25%) members had the replacement of either isoleucine or valine with methionine in YMDD motif. The frequency of mutations from 0 time point to 12 and 24 showed that there was an ascending trend between sequential samples (P values < 0.001). The comparison between groups of II and III showed that mutation was related to valine amino acid (4.5% for 12 months and 11.4% for 24 months) and isoleucine amino acid (9.1% for 12 months and 13.6% for 24 months) replacing for the wild type (methionine). The later finding indicated that the frequency of isoleucine increased as time passed by, but this increase was not statistically significant (P value > 0.05).
| Group I | Group II | Group III | P Value | |
|---|---|---|---|---|
| YMDD mutations | 1 (1.1) | 12 (13.6) | 22 (25.0) | < 0.001 |
| Precore mutations | 31 (35.2) | 36 (41) | 41 (46.6) | < 0.001 |
4.4. Precore Protein
Tables 1 and 2 present the alignment of deduced amino acid sequences of the precore protein obtained from the serial serum samples before administration of LAM and after treatment. In group I, 31 (35.2%); group II, 36 (41.0%), and group III, 41 (46.6%) members had precore stop codon mutations. The frequency of mutations from 0 time point to 12 and 24 showed that there was a rising trend among sequential samples (Figure 1). Indeed, frequency of precore stop codon was significantly increased with the passage of time in the patients of this study (P values < 0.001). Figure 1 shows the pattern of precore and YMDD mutations elevation among these groups of patients.
Prevalence of YIDD and YVDD (Polymerase) and Precore Mutations Patients at Time: 0, 12, and 24 (Months).
5. Discussion
The emergence of drug-resistant variants is one of the most serious problems associated with antiviral therapy. Although the initial effect of the lamivudine in suppressing HBV replication and reducing alanine aminotransferase activity is excellent, the emergence of drug-resistant variants reduces considerably the effect of the drug (29, 30). Precore stop codon mutation is defined as a transition or replacement of G to A at nucleotide position 1896 in precore gene that results in a premature stop codon mutation at translation level, which abrogates the synthesis of HBeAg. Hence, the total loss of HBeAg would mitigate the its suppressive effect on HBV replication, leading to a high viral load profile in HBeAg negative patients who harbor precore stop codon mutations, suggesting that failure to produce a target antigen may be a way to evade the clearance of infected hepatocytes (31). Emergence of this mutation heralds a poor prognosis on the chronicity state of HBV carriers (32). This mutation is common in HBV genotypes B, C, D, and E. A relatively high prevalence of this mutation in the present study was not surprising, as previous reports from Iranian studies showed 38% to 58% of precore mutation (25, 33). Moreover, this is the first Iranian study that investigated the evolution and emergence of this mutation in sequential samples from chronic carriers under antiviral therapy. This study clearly showed that as time went by, the prevalence of precore mutation increased significantly.
Data regarding on the concomitant presence of drug resistance and precore mutants are scarce. One study from Pakistan (34) conducted on 100 chronic carriers on lamivudine therapy, showed a lower prevalence of 6% and 14.3% for YMDD and precore mutations, respectively. After 24 weeks of therapy, 66% of the patients with the latter mutation, seroconverted to anti-HBe and lost their HBV DNA. However, in our study, the rates of precore and YMDD mutations were getting higher with prolonged therapy with no sign of decrease in HBV DNA levels. Whether prolonged lamivudine therapy directly affect the emergence of precore mutations was not clear, as usually a significant proportion of genotype D-infected individuals contain this mutation. On the other hand, the correlation between these two phenomena was significant; indicating that both YMDD and precore mutations, synergically resulted in drug resistance with poor virological response to therapy. Furthermore, the independent impact of precore mutations on the sensitivity for emergence of drug resistant was not clear, as the previous reports from Iranian patients revealed the prevalence of 25.7% (12) to 50% (Jazayeri, unpublished data) of YMDD mutations in patients undergoing lamivudine therapy. We believe that despite these observations, the effects of both mutations on each other are not reciprocal and their emergences may be independent to each one.
In conclusion, the results from this study, confirmed that precore mutations are common amongst Iranian HBV chronic carriers under lamivudine therapy and these mutations are accompanied with clinical relapse. The effect of precore mutation alone on antiviral therapy in these patients is not clear and the exact relationship between the presence of precore mutant strains of HBV and the efficacy of lamivudine therapy needs to be documented by cloning the sequential sera obtained from patients to monitor the emergence, clearance, and probably the re-emergence of precore mutants in a viral quasispecies pool.
Acknowledgements
References
-
1.
Matsuda K. Novel susceptibility loci for hepatocellular carcinoma in chronic HBV carriers. Hepatobiliary Surg Nutr. 2012;1(1):59-60. [PubMed ID: 24570905]. https://doi.org/10.3978/j.issn.2304-3881.2012.10.17.
-
2.
Chen LP, Zhao J, Du Y, Han YF, Su T, Zhang HW, et al. Antiviral treatment to prevent chronic hepatitis B or C-related hepatocellular carcinoma. World J Virol. 2012;1(6):174-83. [PubMed ID: 24175223]. https://doi.org/10.5501/wjv.v1.i6.174.
-
3.
Lai CL, Chien RN, Leung NW, Chang TT, Guan R, Tai DI, et al. A one-year trial of lamivudine for chronic hepatitis B. Asia Hepatitis Lamivudine Study Group. N Engl J Med. 1998;339(2):61-8. [PubMed ID: 9654535]. https://doi.org/10.1056/NEJM199807093390201.
-
4.
Lai CL, Ching CK, Tung AK, Li E, Young J, Hill A, et al. Lamivudine is effective in suppressing hepatitis B virus DNA in Chinese hepatitis B surface antigen carriers: a placebo-controlled trial. Hepatology. 1997;25(1):241-4. [PubMed ID: 8985298]. https://doi.org/10.1002/hep.510250144.
-
5.
Boni C, Penna A, Ogg GS, Bertoletti A, Pilli M, Cavallo C, et al. Lamivudine treatment can overcome cytotoxic T-cell hyporesponsiveness in chronic hepatitis B: new perspectives for immune therapy. Hepatology. 2001;33(4):963-71. [PubMed ID: 11283861]. https://doi.org/10.1053/jhep.2001.23045.
-
6.
Dienstag JL, Schiff ER, Mitchell M, Casey DJ, Gitlin N, Lissoos T, et al. Extended lamivudine retreatment for chronic hepatitis B: maintenance of viral suppression after discontinuation of therapy. Hepatology. 1999;30(4):1082-7. [PubMed ID: 10498663]. https://doi.org/10.1002/hep.510300427.
-
7.
Lai CL, Dienstag J, Schiff E, Leung NW, Atkins M, Hunt C, et al. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin Infect Dis. 2003;36(6):687-96. [PubMed ID: 12627352]. https://doi.org/10.1086/368083.
-
8.
Nafa S, Ahmed S, Tavan D, Pichoud C, Berby F, Stuyver L, et al. Early detection of viral resistance by determination of hepatitis B virus polymerase mutations in patients treated by lamivudine for chronic hepatitis B. Hepatology. 2000;32(5):1078-88. [PubMed ID: 11050059]. https://doi.org/10.1053/jhep.2000.19619.
-
9.
Karayiannis P. Hepatitis B virus: old, new and future approaches to antiviral treatment. J Antimicrob Chemother. 2003;51(4):761-85. [PubMed ID: 12654750]. https://doi.org/10.1093/jac/dkg163.
-
10.
Hosseini SY, Sabahi F, Amini-Bavil-Olyaee S, Alavian SM, Merat S. A novel accurate ACRS-PCR method with a digestion internal control for identification of wild type and YMDD mutants of hepatitis B virus strains. J Virol Methods. 2006;137(2):298-303. [PubMed ID: 16962669]. https://doi.org/10.1016/j.jviromet.2006.07.008.
-
11.
Ghandehari F, Pourazar A, Zadeh MS, Tajedin N. Probing rate of YMDD motif mutant in lamivudine treatment of Iranian patients with chronic hepatitis B virus infection. Asian J Transfus Sci. 2011;5(1):32-4. [PubMed ID: 21572712]. https://doi.org/10.4103/0973-6247.75982.
-
12.
Afshar RM, Mollaie HR. Detection of HBV resistance to lamivudine in patients with chronic hepatitis B using Zip nucleic acid probes in Kerman, southeast of Iran. Asian Pac J Cancer Prev. 2012;13(8):3657-61. [PubMed ID: 23098450].
-
13.
Carman WF, Jacyna MR, Hadziyannis S, Karayiannis P, McGarvey MJ, Makris A, et al. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;2(8663):588-91. [PubMed ID: 2570285].
-
14.
Gandhe SS, Chadha MS, Walimbe AM, Arankalle VA. Hepatitis B virus: prevalence of precore/core promoter mutants in different clinical categories of Indian patients. J Viral Hepat. 2003;10(5):367-82. [PubMed ID: 12969189].
-
15.
Hasegawa K, Huang J, Rogers SA, Blum HE, Liang TJ. Enhanced replication of a hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. J Virol. 1994;68(3):1651-9. [PubMed ID: 8107226].
-
16.
Friedt M, Gerner P, Lausch E, Trubel H, Zabel B, Wirth S. Mutations in the basic core promotor and the precore region of hepatitis B virus and their selection in children with fulminant and chronic hepatitis B. Hepatology. 1999;29(4):1252-8. [PubMed ID: 10094972]. https://doi.org/10.1002/hep.510290418.
-
17.
Takahashi K, Akahane Y, Hino K, Ohta Y, Mishiro S. Hepatitis B virus genomic sequence in the circulation of hepatocellular carcinoma patients: comparative analysis of 40 full-length isolates. Arch Virol. 1998;143(12):2313-26. [PubMed ID: 9930189].
-
18.
Zhong S, Chan JY, Yeo W, Tam JS, Johnson PJ. Frequent integration of precore/core mutants of hepatitis B virus in human hepatocellular carcinoma tissues. J Viral Hepat. 2000;7(2):115-23. [PubMed ID: 10760041].
-
19.
Gunther S, Meisel H, Reip A, Miska S, Kruger DH, Will H. Frequent and rapid emergence of mutated pre-C sequences in HBV from e-antigen positive carriers who seroconvert to anti-HBe during interferon treatment. Virology. 1992;187(1):271-9. [PubMed ID: 1736529].
-
20.
Zhang X, Zoulim F, Habersetzer F, Xiong S, Trepo C. Analysis of hepatitis B virus genotypes and pre-core region variability during interferon treatment of HBe antigen negative chronic hepatitis B. J Med Virol. 1996;48(1):8-16. [PubMed ID: 8825704]. https://doi.org/10.1002/(SICI)1096-9071(199601)48:1<8::AID-JMV2>3.0.CO;2-E.
-
21.
Lee YI, Hur GM, Suh DJ, Kim SH. Novel pre-C/C gene mutants of hepatitis B virus in chronic active hepatitis: naturally occurring escape mutants. J Gen Virol. 1996;77 ( Pt 6):1129-38. [PubMed ID: 8683197].
-
22.
Laras A, Koskinas J, Hadziyannis SJ. In vivo suppression of precore mRNA synthesis is associated with mutations in the hepatitis B virus core promoter. Virology. 2002;295(1):86-96. [PubMed ID: 12033768]. https://doi.org/10.1006/viro.2001.1352.
-
23.
Veazjalali M, Norder H, Magnius L, Jazayeri SM, Alavian SM, Mokhtari-Azad T. A new core promoter mutation and premature stop codon in the S gene in HBV strains from Iranian patients with cirrhosis. J Viral Hepat. 2009;16(4):259-64. [PubMed ID: 19222745]. https://doi.org/10.1111/j.1365-2893.2009.01069.x.
-
24.
Poustchi H, Mohamadkhani A, Bowden S, Montazeri G, Ayres A, Revill P, et al. Clinical significance of precore and core promoter mutations in genotype D hepatitis B-related chronic liver disease. J Viral Hepat. 2008;15(10):753-60. [PubMed ID: 18507754]. https://doi.org/10.1111/j.1365-2893.2008.00998.x.
-
25.
Bahramali G, Sadeghizadeh M, Amini-Bavil-Olyaee S, Alavian SM, Behzad-Behbahani A, Adeli A, et al. Clinical, virologic and phylogenetic features of hepatitis B infection in Iranian patients. World J Gastroenterol. 2008;14(35):5448-53. [PubMed ID: 18803358].
-
26.
Mahabadi M, Norouzi M, Alavian SM, Samimirad K, Azad TM, Saberfar E, et al. Drug-related mutational patterns in hepatitis B virus (HBV) reverse transcriptase proteins from Iranian treatment-naive chronic HBV patients. Hepat Mon. 2013;13(1). eee6712. [PubMed ID: 23596461]. https://doi.org/10.5812/hepatmon.6712.
-
27.
Jazayeri SM, Basuni AA, Sran N, Gish R, Cooksley G, Locarnini S, et al. HBV core sequence: definition of genotype-specific variability and correlation with geographical origin. J Viral Hepat. 2004;11(6):488-501. [PubMed ID: 15500549]. https://doi.org/10.1111/j.1365-2893.2004.00534.x.
-
28.
Khedive A, Norouzi M, Ramezani F, Karimzadeh H, Alavian SM, Malekzadeh R, et al. Hepatitis B virus surface protein mutations clustered mainly in CTL immune epitopes in chronic carriers: results of an Iranian nationwide study. J Viral Hepat. 2013;20(7):494-501. [PubMed ID: 23730843]. https://doi.org/10.1111/jvh.12045.
-
29.
Chayama K, Suzuki Y, Kobayashi M, Kobayashi M, Tsubota A, Hashimoto M, et al. Emergence and takeover of YMDD motif mutant hepatitis B virus during long-term lamivudine therapy and re-takeover by wild type after cessation of therapy. Hepatology. 1998;27(6):1711-6. [PubMed ID: 9620347]. https://doi.org/10.1002/hep.510270634.
-
30.
Liaw YF, Chien RN, Yeh CT, Tsai SL, Chu CM. Acute exacerbation and hepatitis B virus clearance after emergence of YMDD motif mutation during lamivudine therapy. Hepatology. 1999;30(2):567-72. [PubMed ID: 10421670]. https://doi.org/10.1002/hep.510300221.
-
31.
Carman W, Thomas H, Domingo E. Viral genetic variation: hepatitis B virus as a clinical example. Lancet. 1993;341(8841):349-53. [PubMed ID: 8094122].
-
32.
Papatheodoridis GV, Hadziyannis SJ. Diagnosis and management of pre-core mutant chronic hepatitis B. J Viral Hepat. 2001;8(5):311-21. [PubMed ID: 11555188].
-
33.
Amini-Bavil-Olyaee S, Sarrami-Forooshani R, Adeli A, Mahboudi F, Sabahi F, Nafisi H, et al. A novel accurate amplification created restriction site method for determination of the wild type and the precore mutant hepatitis B virus variants. J Virol Methods. 2005;127(1):19-23. [PubMed ID: 15893561]. https://doi.org/10.1016/j.jviromet.2005.02.011.
-
34.
Qureshi H, Arif A, Ahmed W, Alam SE. YMDD mutation in Pakistani patients. The comparison of Eastern response with the Western response. J Pak Med Assoc. 2009;59(12):858-9. [PubMed ID: 20201183].