Abstract
Background:
Methicillin-Resistant Staphylococcus aureus (MRSA) is a major pathogen in the hospital and community settings. Rapid methods to diagnose S. aureus infections are sought by many researchers worldwide. The current study aimed to utilize a phenotypic method of turanose fermentation to identify methicillin-susceptible and resistant S. aureus.Objectives:
The current study aimed to assay the turanose metabolism at different dilutions as a rapid phenotypic method to identify MRSA isolates.Materials and Methods:
A total of 150 Staphylococcus isolates were collected from Tehran health centers. Staphylococcus aureus isolates were identified based on cultural characteristics, biochemical reactions and positive tube coagulase test. Methicillin resistance was determined by the disk diffusion method. The Polymerase Chain Reaction amplification was used to detect the mecA gene in MRSA isolates. All the methicillin-resistant and susceptible isolates were evaluated for turanose metabolism with 1%, 0.7% and 0.5% dilutions using the microplate method.Results:
Out of the 150 staphylococcal isolates, 80 were identified as S. aureus. Among which 40 (50%) of the isolates were MRSA. The mecA gene was present in all S. aureus isolates resistant to methicillin. A considerable difference was also observed between susceptible and resistant isolates of S. aureus at a 0.7% dilution of turanose.Conclusions:
Since it is highly important to rapidly detect MRSA isolates, especially in nosocomial infections, phenotypic methods may certainly be useful for this purpose. Resistance to methicillin in S. aureus shows a substantially increased ability in turanose metabolism. It is concluded that fermentation of turanose at 0.7% dilution could be a rapid detection method for primary screening of MRSA isolates.Keywords
1. Background
Staphylococcus aureus is an important bacterial pathogen that may cause a variety of diseases, ranging from common skin and soft tissue infections to very serious and lethal diseases such as septic shock, toxic shock syndrome or pneumonia (1, 2). Although antibiotic therapy is currently used to treat S. aureus infections, the emergence of antibiotic-resistant strains is rapidly exhausting available treatment options (1, 2). To date, resistance of S. aureus strains to methicillin is categorized as one the main causes of hospital infections. Furthermore, treatment of methicillin-resistant S. aureus (MRSA) infections often requires longer hospital stays and imposes a tremendous financial burden (3). Severe MRSA infections therapy is complicated by the fact that these strains are susceptible to only a few antimicrobials such as vancomycin. Unfortunately, in the past decade, vancomycin resistant S. aureus is isolated, highlighting the ability of the bacteria to adapt to the new antibiotics (4, 5).
The main resistance mechanism of S. aureus to methicillin is the acquisition of the mecA gene and encoding a new transpeptidase enzyme (PBP) that has a reduced tendency to beta lactam antibiotics. Since mecA gene has high homology among MRSA strains and coagulase negative Staphylococcus resistant to methicillin, it is considered as a proper molecular marker to determine methicillin resistance in all Staphylococcal species (6, 7). Frequency of the mecA gene among MRSA strains varies in different countries (8). These differences appear to be due to various distribution of this gene at different geographical regions or their identification method. Increased spread of mecA is the common issue among all these studies, indicating a potential threat that make staphylococcal infections resistant to methicillin and other globally used antibiotics (9).
Nowadays, phenotypic methods such as disk diffusion and minimum inhibitory concentration (MIC), as well as genotypic methods such as PCR are used to diagnose MRSA strains. These methods have some limitations and there is no perfect method to diagnose these strains (10). As mentioned above, since MRSA strains are among the important human pathogens, it is necessary to find a reliable and simple method as an alternative or complementary method to diagnose MRSA strains especially during the prevalence of the strains or performance of a health plan.
Turanose is a reducing disaccharide and an isomer of sucrose (11). Sucrose is used as a carbon source by many bacteria and fungi. For instance, sucrose is used by almost 98% of S. aureus strains, producing acid during its fermentation process. It is used to diagnose these strains (12). The current study assayed the turanose metabolism at different dilutions as a new rapid phenotypic method to identify MRSA isolates. The obtained results indicated a considerable difference between the fermentation of turanose in MRSA and methicillin-sensitive S. aureus (MSSA) isolates at 0.7% dilution. Therefore, a simple phenotypic test based on the turanose fermentation is suggested as a useful method to diagnose MRSA strains.
2. Objectives
Due to the importance of methicillin-resistant S. aureus (MRSA) infections, the current study aimed to assay the turanose metabolism at different dilutions as a rapid phenotypic method to identify MRSA isolates.
3. Materials and Methods
3.1. Sample Collection
Samples were collected from Tehran medical centers, within three months (October-December 2011). Isolation of S. aureus was performed according to the standard microbial and biochemical tests, such as fermentation of mannitol, coagulase test and DNase (Merck, KGaA, Darmstadt, Germany). S. aureus strains were considered positive for the tests (12).
3.2. Disk Diffusion Test
The disk diffusion method was performed using a disk containing 1 μg oxacillin (BBL, Sensi Disk, USA) to diagnose MRSA isolates (12). This method was performed according to the Clinical and laboratory standards institute (CLSI) guidelines (13). For this purpose, bacterial suspensions equal to 0.5 McFarland standard were scattered on agar Mueller-Hinton medium (Merck, KGaA, Darmstadt, Germany). Then, the oxacillin disk was put on the incubated plates and after 24 hours the inhibition zone diameter around the disk was measured.
3.3. Detection of mecA gene by Polymerase Chain Reaction
DNA extraction of S. aureus isolates was conducted by DNA extraction kit (Qiagen GmbH, Hilden, Germany) to identify the presence or absence of mecA gene in the S. aureus isolates, the following pairs of primers were used for the amplification (14):
MecA F: 5'-TAGAAATGACTGAACGTCCGATAA-3'
MecA R: 5'-CCAATTCCATGTTTCGGTCTAA-3'
PCR reactions were carried out in a final volume of 50 μL containing 2 μL of DNA template, 5 μL of 10x reaction buffer, 2 μL of dNTPs (10 mM), 2 μL of Mgcl2 (50 mM), 2 μL of each primer (10 pmol), and 1U of Taq DNA polymerase (Fermentas; Vilnius, Lithuania). The PCR conditions were as follow: an initial denaturation for 5 minutes at 94°C, followed by 30 cycles, each consisting of 60 seconds at 94°C, 60 seconds at 55°C, and 60 seconds at 72°C with a final extension step at 72°C for 5 minutes. After amplification of mecA gene, 10 μL of the samples were subjected to electrophoresis on a 1% agarose gel to confirm the presence of the amplified products.
3.4. Metabolism of Turanose
Turanose was purchased from Merck company. Five grams of turanose was dissolved in 100 mL of distilled water and sugar with 5% dilution was prepared. To complete tantalization process of the turanose, the mixture was placed in a boiling water bath (80°C) for sterilization, for an hour per day on three consecutive days. The prepared 5% dilution of turanose was used to provide 1%, 0.7% and 0.5% dilutions. Preparation of microbial suspensions of each S. aureus strain was performed in sterile glass vials and their turbidity was adjusted to 0.5 McFarland. In triplicate, 150 μL of nutrient broth (sugar free) medium containing different turanose dilutions (0.5%, 0.7% and 1%) were inoculated to each well in microplates (Greiner, Germany) and 150 μL of each microbial suspension was added to the microplates. Furthermore, medium as well as turanose without microbial suspension were used as a negative control and blank, respectively. Then, the inoculated microplates were put into incubator (37°C) overnight. The growth of susceptible and resistant isolates to methicillin was assayed in the presence of different dilutions of turanose by measurement of their light absorption with enzyme-linked immunosorbent assay (ELISA) reader (Awareness Technology Inc.) at 610 nm. To calculate the average, the adsorption of the isolates was read three times.
3.5. Statistical Analysis
One-way ANOVA, Student᾿s t-test and Tukey HSD test were used to compare the differences between the mean values of the groups using SPSS software. P < 0.05 of all results was considered as significant.
4. Results
4.1. Sample Identification
Out of the 150 Staphylococcus isolates, 80 were identified as S. aureus. According to the disk diffusion results, 40 S. aureus isolates were resistant-to-methicillin and 40 were susceptible.
4.2. Detection of MRSA Isolates
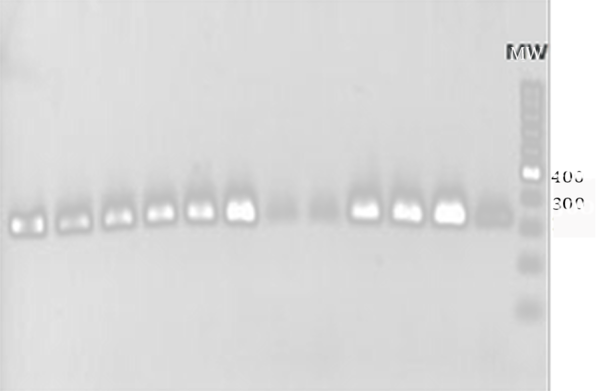
All of the MRSA isolates diagnosed with the disk diffusion method contained mecA gene. Electrophoresis of PCR products showed that the length of PCR fragment of the mecA gene was approximately 310 bp (Figure 1).
PCR Product of mecA Gene in the MSRA Isolates
4.3. Metabolism of Turanose in MRSA and MSSA Isolates
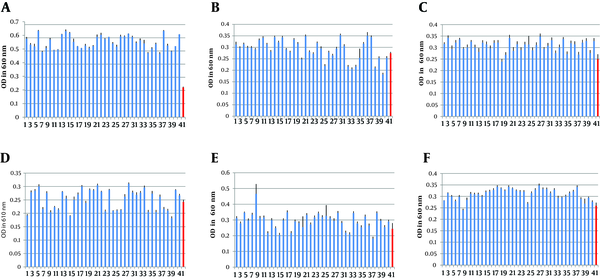
The growth of MRSA and MSSA strains was evaluated in the presence of turanose with 0.5%, 0.7% and 1% dilutions to find significant differences in metabolism of turanose among the strains. The result of turanose metabolism at the dilutions showed significant difference among MRSA and MSSA isolates in 0.7% dilution of turanose (P < 0.003) (Figure 2). However, the differences between MRSA and MSSA isolates in 1% and 0.5% dilutions were not statistically significant (P > 0.05) (Figure 2).
Fermentation of Turanose by the MRSA and MSSA Strains in 0.7%, 1% and 0.5% Dilutions.
5. Discussion
Staphylococcus aureus is one of the major causes of nosocomial and community-acquired infections that results in substantial morbidity and mortality (1, 2). Furthermore, due to multi-drug resistance and the ability to acquire resistance to new antibiotics quickly, it is more and more difficult to treat S. aureus infection, especially with the emergence of vancomycin resistant S. aureus (15, 16). After the growth of MRSA at hospitals, these infections gradually go out of hospitals, which worsen their treatment complexity (4). The prevalence of MRSA varies in different countries. For example, the prevalence of MRSA was reported 2% in Switzerland and the Netherlands or 80% in China (17). There were some studies on MRSA prevalence in Iran. Similarly, high prevalence of MRSA was reported in different parts of Iran (18, 19). The differences in prevalence of the MRSA strains may be due to long-term antibiotic treatment of patients suffering from severe infections, with long hospitalization and increased selective pressure (20).
The mecA gene is considered a reserved indicator in MRSA strains. As a result, mecA gene is detected by PCR in most studies to confirm phenotypic tests in MRSA diagnosis (20). In the current study, all S. aureus strains resistant to oxacillin in disk diffusion represented the mecA gene. Rodriguez reported that all MRSA strains contain the mecA gene, similar to the current study. However, there are reports on the absence of this gene in MRSA strains. In a study, it was shown that 2.25% of MRSA strains lack mecA gene (12). This discrepancy may be due to bad quality of the PCR test or genetic mutation. Different methods are offered to diagnose MRSA strains (10). Although, genotypic methods such as PCR are in the category of rapid methods to diagnose resistant strains, they are not perfect enough to diagnose resistance. For example, whereas detection of the mecA gene is the gold standard of identification of MRSA strains, the required facilities and high costs do not let many laboratories use the method to identify MRSA (21). Furthermore, it is documented that some resistant strains have different genetic sequences and the designed primers fail to identify the MRSA strains in PCR reaction. This difference in genetic sequence indicates that the necessary precaution should be noted in the application of genotypic methods to detect MRSA. In addition to being time-consuming and expensive, phenotypic methods such as disk diffusion have difficulties to diagnose resistant strains (22).
Despite extensive efforts, a diagnostic method identifying all MRSA is currently missing and there is a need to make future diagnostic test evaluations to develop a sensitive, specific and practical method to detect the strains (22). Phenotypic diagnostic methods of bacteria are on the basis of quantitative assessment of products obtained from biochemical reactions or metabolism of initial substrates. Since biochemical and phenotypic methods completely depend on responsible genes on the bacterial DNA, these methods are a kind of indirect assessment of bacterial genetics. Although, phenotypic methods are usually simpler than genotypic methods, genotypic methods appear to be necessary in some cases in order to confirm phenotypic findings (9, 10).
The current study was designed based on the kinetic assessment of biochemical reactions. Some studies showed that genes are responsible for resistance to methicillin make some changes in the walls of Gram-positive bacteria and there is a relationship between the wall changes made by resistance genes and metabolism of turanose in MRSA strains (23). Thus, the relationship between metabolism of turanose and resistance to methicillin in S. aureus strains was evaluated in the current study. The results showed a significant relationship between the metabolism of turanose at 0.7% dilution and resistance to methicillin in S. aureus strains. By turanose metabolism as a phenotypic test, it was found that a simple diagnosis method can be achieved to identify these strains. Therefore, such studies that can lead to find a rapid way to diagnose antibiotic resistance is highly valuable. It is likely to introduce rapid diagnostic kits of these bacteria by analyzing bigger populations of the strains.
In conclusion, the current study suggests a new method for rapid and simple diagnosis of MRSA strains at laboratories by measuring the metabolism of turanose.
Acknowledgements
References
-
1.
Brady RA, Mocca CP, Prabhakara R, Plaut RD, Shirtliff ME, Merkel TJ, et al. Evaluation of genetically inactivated alpha toxin for protection in multiple mouse models of Staphylococcus aureus infection. PLoS One. 2013;8(4). eee63040. [PubMed ID: 23658662]. https://doi.org/10.1371/journal.pone.0063040.
-
2.
Schaffer AC, Lee JC. Vaccination and passive immunisation against Staphylococcus aureus. Int J Antimicrob Agents. 2008;32 Suppl 1:S71-8. [PubMed ID: 18757184]. https://doi.org/10.1016/j.ijantimicag.2008.06.009.
-
3.
Malani PN, Thal L, Donabedian SM, Robinson-Dunn B, Kauffman CA, Chow JW, et al. Molecular analysis of vancomycin-resistant Enterococcus faecalis from Michigan hospitals during a 10 year period. J Antimicrob Chemother. 2002;49(5):841-3. [PubMed ID: 12003981].
-
4.
Kim HK, Thammavongsa V, Schneewind O, Missiakas D. Recurrent infections and immune evasion strategies of Staphylococcus aureus. Curr Opin Microbiol. 2012;15(1):92-9. [PubMed ID: 22088393]. https://doi.org/10.1016/j.mib.2011.10.012.
-
5.
Hu DL, Narita K, Hyodo M, Hayakawa Y, Nakane A, Karaolis DK. c-di-GMP as a vaccine adjuvant enhances protection against systemic methicillin-resistant Staphylococcus aureus (MRSA) infection. Vaccine. 2009;27(35):4867-73. [PubMed ID: 19406185]. https://doi.org/10.1016/j.vaccine.2009.04.053.
-
6.
Ardic N, Sareyyupoglu B, Ozyurt M, Haznedaroglu T, Ilga U. Investigation of aminoglycoside modifying enzyme genes in methicillin-resistant staphylococci. Microbiol Res. 2006;161(1):49-54. [PubMed ID: 16338590]. https://doi.org/10.1016/j.micres.2005.05.002.
-
7.
Martins A, Cunha Mde L. Methicillin resistance in Staphylococcus aureus and coagulase-negative staphylococci: epidemiological and molecular aspects. Microbiol Immunol. 2007;51(9):787-95. [PubMed ID: 17895595].
-
8.
Diekema DJ, Pfaller MA, Schmitz FJ, Smayevsky J, Bell J, Jones RN, et al. Survey of infections due to Staphylococcus species: frequency of occurrence and antimicrobial susceptibility of isolates collected in the United States, Canada, Latin America, Europe, and the Western Pacific region for the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin Infect Dis. 2001;32 Suppl 2:S114-32. [PubMed ID: 11320452]. https://doi.org/10.1086/320184.
-
9.
Wallet F, Roussel-Delvallez M, Courcol RJ. Choice of a routine method for detecting methicillin-resistance in staphylococci. J Antimicrob Chemother. 1996;37(5):901-9. [PubMed ID: 8737140].
-
10.
Archer GL, Niemeyer DM. Origin and evolution of DNA associated with resistance to methicillin in staphylococci. Trends Microbiol. 1994;2(10):343-7. https://doi.org/10.1016/0966-842X(94)90608-4.
-
11.
Alexander D, Hoffmann J, Munz A, Friedrich B, Geis-Gerstorfer J, Reinert S. Comparison of three dimensional scaffolds for bone engineering constructs using human jaw periosteal cells. J Stem Cells Regen Med. 2007;2(1):177. [PubMed ID: 24692984].
-
12.
Hiramatsu K, Cui L, Kuroda M, Ito T. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 2001;9(10):486-93. [PubMed ID: 11597450].
-
13.
Performance standards for antimicrobial disc diffusion tests Approved standards. Wayne: CLSI; 2006.
-
14.
Chaieb K, Touati A, Salah AM, Hassen AB, Mahdouani K, Bakhrouf A. DNA fingerprinting of a multi-resistant coagulase-negative staphylococci isolated from biomaterials in dialysis services. Arch Med Res. 2006;37(8):953-60. [PubMed ID: 17045110]. https://doi.org/10.1016/j.arcmed.2006.06.007.
-
15.
Kuklin NA, Clark DJ, Secore S, Cook J, Cope LD, McNeely T, et al. A novel Staphylococcus aureus vaccine: iron surface determinant B induces rapid antibody responses in rhesus macaques and specific increased survival in a murine S. aureus sepsis model. Infect Immun. 2006;74(4):2215-23. [PubMed ID: 16552052]. https://doi.org/10.1128/IAI.74.4.2215-2223.2006.
-
16.
Proctor RA. Challenges for a universal Staphylococcus aureus vaccine. Clin Infect Dis. 2012;54(8):1179-86. [PubMed ID: 22354924]. https://doi.org/10.1093/cid/cis033.
-
17.
Wisplinghoff H, Ewertz B, Wisplinghoff S, Stefanik D, Plum G, Perdreau-Remington F, et al. Molecular evolution of methicillin-resistant Staphylococcus aureus in the metropolitan area of Cologne, Germany, from 1984 to 1998. J Clin Microbiol. 2005;43(11):5445-51. [PubMed ID: 16272468]. https://doi.org/10.1128/JCM.43.11.5445-5451.2005.
-
18.
Fatholahzadeh B, Emaneini M, Aligholi M, Gilbert G, Taherikalani M, Jonaidi N, et al. Molecular characterization of methicillin-resistant Staphylococcus aureus clones from a teaching hospital in Tehran. Jpn J Infect Dis. 2009;62(4):309-11. [PubMed ID: 19628913].
-
19.
Nikbakht M, Nahaei MR, Akhi MT, Asgharzadeh M, Nikvash S. Molecular fingerprinting of meticillin-resistant Staphylococcus aureus strains isolated from patients and staff of two Iranian hospitals. J Hosp Infect. 2008;69(1):46-55. [PubMed ID: 18358563]. https://doi.org/10.1016/j.jhin.2008.01.034.
-
20.
Graham JC, Gould FK. Role of aminoglycosides in the treatment of bacterial endocarditis. Antimicrob Chemother. 2002;49(3):437-44.
-
21.
Mahbub Alam M, Kobayashi N, Ishino M, Sumi A, Kobayashi K, Uehara N, et al. Detection of a novel aph(2") allele (aph[2"]-Ie) conferring high-level gentamicin resistance and a spectinomycin resistance gene ant(9)-Ia (aad 9) in clinical isolates of enterococci. Microb Drug Resist. 2005;11(3):239-47. [PubMed ID: 16201926]. https://doi.org/10.1089/mdr.2005.11.239.
-
22.
Agvald-Ohman C, Wernerman J, Nord CE, Edlund C. Anaerobic bacteria commonly colonize the lower airways of intubated ICU patients. Clin Microbiol Infect. 2003;9(5):397-405. [PubMed ID: 12848752].
-
23.
Perez-Roth E, Claverie-Martin F, Villar J, Mendez-Alvarez S. Multiplex PCR for simultaneous identification of Staphylococcus aureus and detection of methicillin and mupirocin resistance. J Clin Microbiol. 2001;39(11):4037-41. [PubMed ID: 11682527]. https://doi.org/10.1128/JCM.39.11.4037-4041.2001.