Abstract
Background:
Staphylococcus aureus, an important human pathogen is one of the main causative agents of nosocomial infection. Virulence genes play a major role in the pathogenicity of this agent and its infections. Methicillin-Resistant Staphylococcus aureus (MRSA) isolates are major challenge among infectious agents that can cause severe infections and mortality. Methicillin-resistant S. aureus produces a unique type of Penicillin Binding Protein 2a (PBP2a) that has low affinity for β-lactam antibiotics. Most of the MRSA bacterial strains can also produce a leukotoxin as Panton-Valentine Leukocidin (PVL) that increases the virulence of MRSA strains and can cause severe necrotic pneumonia. The presence of pvl gene is a genetic marker for the MRSA populations.Objectives:
The aim of this study was to explore the association of pvl and mecA genes in clinical isolates of MRSA.Materials and Methods:
Fifty MRSA isolates were collected from 200 clinical samples from three different educational hospitals in Ahvaz, Iran, and identified by biochemical tests including catalase, oxidase, tube coagulase, mannitol fermentation, and sensitivity to furazolidone, resistance to bacitracin, PYR test and Voges-Proskauer test. Their resistance to methicillin was evaluated using the disc diffusion method. DNA was extracted by boiling and then the presence of pvl and mecA genes was investigated by the polymerase chain reaction method using specific primers.Results:
The results revealed that mecA and pvl genes were positive for 15 (30%) and 3 (6%) of the isolates, respectively. None of mecA positive isolates was positive for pvl gene.Conclusions:
It can be concluded from these results that fortunately the prevalence of pvl gene is low in MRSA isolates in this region and there is no association between the presence of pvl and mecA genes in these isolates.Keywords
MRSA Iran Virulence Factor Methicillin Resistance Panton Valentine Leukocidin
1. Background
Among the etiologic agents of infectious disease, Staphylococcus aureus, especially Methicillin-Resistant Staphylococcus aureus (MRSA) strains are the main agents of human infections (1). This pathogen can cause nosocomial infection in such a manner that it is the most common pathogen isolated from patients in hospitals that can cause wide range of infections from skin and soft tissue infections till toxic shock syndrome and life threatening syndromes. During the end of 1990s, increasing of S. aureus infections was led to isolation of MRSA (2-5). Nearly, all S. aureus isolates produce various toxins and enzymes that are important in the pathogenicity of this bacterium (6, 7). With the emergence of resistant strains at 1961 that had multidrug resistance, the treatment of S. aureus infections had been paid more attention. The MRSA stains are widely distributed around the world that shows their resistance to both penicillins and cephalosporins (8, 9).
The MRSA produces a unique type of Penicillin Binding Protein 2a (PBP2a) that has low affinity for β-lactam antibiotics (10, 11). Most of MRSA strains can also produce a leukotoxin as Panton-Valentine Leukocidin (PVL) that increases their virulence and can cause severe necrotic pneumonia (8, 12). Pathogenicity of S. aureus results from the surface cell wall structures and different exoproteins that PVL is one of the most important of them for overcoming host immunity and disease progress (13). Pore forming toxins perforate membranes of host cells, mainly the plasma membrane and also intracellular organelle membranes. They directly kill target cells, in order to intracellular delivery of other bacterial or external factors, releasing nutrients or escaping from phagosomal space. Panton-valentine leukocidin is a member of pore forming toxins that targets host leukocytes. The PVL- producing strains are commonly involved in the skin and soft tissue infections as well as able to cause life threatening infections like pneumonia (7, 8, 12). Two open reading frames are responsible for coding PVL, i.e. lukS-pv and lukF-pv. The presence of pvl gene is a genetic marker for the MRSA populations (14). Recently, reports about infections caused by pvl-positive strains have been increased that necessitate investigations about the prevalence of this virulent marker among MRSA strains in hospital and also community acquired infections.
2. Objectives
The aim of the present study was to discover the association of pvl and mecA in MRSA strains isolated from hospitalized patients in three educational hospitals of Ahvaz City, Iran.
3. Materials and Methods
3.1. Sampling
Fifty S. aureus isolates were collected from patients in three educational hospitals of Ahvaz, including Imam Khomeini, Golestan and Taleghni hospitals. These samples were obtained from 200 samples collected from different sections of the above-mentioned hospitals including Outpatient Department (OPD), skin, men surgery, ophthalmology, nephrology, pediatric, women, orthopedic, ear, nose and throat (ENT), neonates and internal medicine. Samples were collected from skin lesions, blood cultures, burns, intravenous catheters, wound drainages, abscesses, tracheal secretions, synovial fluids, ocular secretions and urines. These isolates were identified based on standard morphological and biochemical tests including catalse, oxidase, tube coagulase, mannitol fermentation, sensitivity to furazolidon, resistance to bacitracine, PYR test and Voges-Proskauer (15).
3.2. Determining Antibiotic Susceptibility
In order to determine methicillin resistance of isolates Muller-Hinton Agar (MHA, Merck, Germany) screening test was used according to CLSI (clinical and laboratory standards institute) (16). Briefly, a 0.5 McFarland suspension was prepared from pure culture of isolates and then cultured on MHA containing oxacillin (6 μg/mL). For this purpose using a sterile swab the bacterium was inoculated as a dot with a 10 - 15 mm diameter and a strike culture was prepared on the other part of plate and incubated at 35°C. The results were studied after 24 and 48 hours of incubation. Growth of only one colony on this culture medium revealed methicillin resistance of that isolate. Staphylococcus aureus ATCC 29213 as the negative control and sensitive to methicillin and S. aureus ATCC 33591 as the positive control and resistant to methicillin were used in all experiments. Furthermore, the susceptibility of isolates to oxacillin was surveyed using the standard Kirby-Bauer disc diffusion method (17).
3.3. DNA Extraction
The isolates were cultured in Trypticase Soy Broth (TSB, Merck, Germany) for 18 h at 37°C. Then, 1.5 mL of culture was centrifuged (12000 rpm, 10 minutes) and the precipitate was dissolved in one mL deionized water and boiled for 20 minutes. After centrifugation (5000 rpm, 5 minutes) the supernatant was slowly mixed with phenol-chloroform-isoamyl alcohol (1: 24: 25) at 1: 1 ratio and the aqueous phase was harvested by centrifugation at 13000 rpm, 30 seconds. Cold isopropanol was added to aqueous phase (60.100, v/v) and stored overnight at -22°C. The DNA was then precipitated and washed subsequently with absolute and 70% ethanol. Finally, DNA was air-dried at 37°C and dissolved in 200 μL sterile deionized water and stored at -22°C till experiments (18).
3.4. Polymerase Chain Reaction
Polymerase Chain Reaction (PCR) was performed in a final 25 μL volume reaction containing PCR buffer (10x), Magnesium Chloride (MgCl2) (2 mM), Deoxynucleotide Triphosphates (dNTPs) (0.2 mM), forward and reverse primers (10 ρmol/μL), template DNA (1 μL), Taq DNA polymerase ( 1.5 u) and deionized water. The PCR method was performed in thermal cycler (Bio-Rad, USA) according to the following program: initial denaturation (94°C, 2 minutes), 35 cycles each composed of initial denaturation (94°C, 30 seconds), primer annealing (55°C, 45 seconds) and extension (72°C, 75 seconds) and a final extension (72°C, 4 minutes) (19). Positive control for mecA and pvl genes and negative control (distilled water) were also regarded in each series of PCR reaction. The PCR product was subjected to electrophoresis in 1% agarose gel containing DNA safe stain and documented using gel documentation (UVI TEC, Cambridge, UK).
Specific Primers for Amplification of mecA and pvl Genes of Staphylococcus aureus
| Primer | Sequence | Base |
|---|---|---|
| MecA1a | 5´GTAGAAATACT GAACGT CCGATAA 3´ | 24 |
| MecA2 | 5´CCAATTCCACATTGTTTCGGTCTAA 3´ | 25 |
| Luk- PV1 | 5´ATCATTAGGTAAAATGTCTGGACATGATCCA3´ | 31 |
| Luk- PV2 | 5´GCATCAAGTGTATTGGATAGCAAA AGC 3´ | 27 |
4. Results
From a total of 50 samples, 27 samples (54%) were belonged to Imam Khomeini Hospital, 16 samples (32%) from Golestan Hospital and 7 samples (14%) from Taleghani Hospital. According to the patient gender, 26 samples (52%) were isolated from men while 24 samples (48%) were from women. The number of samples from different clinical specimens and hospital sections are presented in Tables 2 and 3, respectively.
The Source of Staphylococcus aureus Isolates in Clinical Samples
| Clinical Sample | Number of Isolates |
|---|---|
| Skin lesions | 11 |
| Blood cultures | 9 |
| Burns | 6 |
| Urines | 3 |
| Abscesses | 3 |
| Tracheal Secretions | 3 |
| Synovial Fluids | 2 |
| Ocular Secretions | 3 |
| Intravenous Catheters | 6 |
| Wound Drainages | 4 |
| Ward | Number of Isolates |
|---|---|
| OPD | 13 |
| Skin | 13 |
| Men surgery | 1 |
| Ophthalmology | 3 |
| Nephrology | 8 |
| Pediatric | 3 |
| Women | 3 |
| Orthopedic | 2 |
| ENT | 2 |
| Neonates | 1 |
| Internal Medicine | 1 |
The results of antibiotic susceptibility tests revealed that all isolates were resistant to methicillin. Following PCR reaction with mecA and pvl specific primers, 314 bp and 433 bp PCR products were produced for mecA and pvl genes, respectively (Figure 1).
Analysis of mecA (314 bp) and pvl (433 bp) Amplification
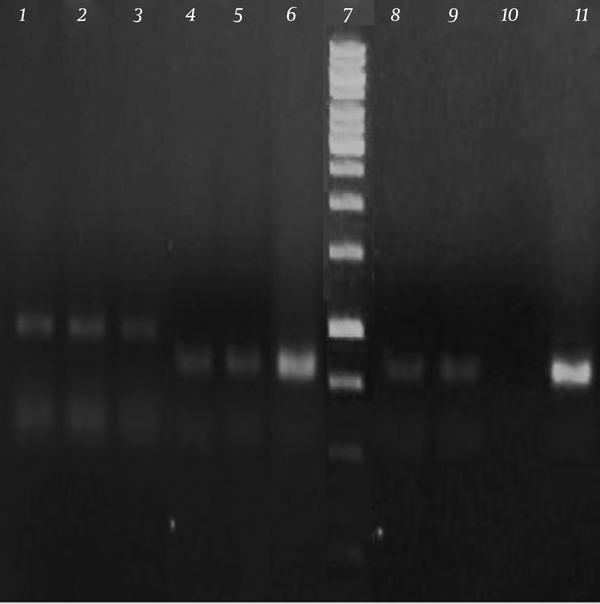
Table 4 shows the total number of MRSA isolates and their mecA and pvl markers. Based on the obtained results, 30% (15 samples) of MRSA strains were positive for mecA gene while 6% (3 samples) had pvl gene. None of the samples had simultaneously both virulence markers.
Results of mecA and pvl Screening in 50 Methicillin-Resistant Staphylococcus aureus Isolates
| Isolate | mecA | pvl |
|---|---|---|
| 1 | - | - |
| 2 | - | - |
| 3 | - | - |
| 4 | - | - |
| 5 | - | - |
| 6 | - | - |
| 7 | - | - |
| 8 | + | - |
| 9 | + | - |
| 10 | - | - |
| 11 | - | - |
| 12 | - | + |
| 13 | + | - |
| 14 | - | - |
| 15 | - | - |
| 16 | - | - |
| 17 | - | - |
| 18 | + | - |
| 19 | - | - |
| 20 | - | - |
| 21 | + | - |
| 22 | - | - |
| 23 | - | - |
| 24 | + | - |
| 25 | - | - |
| 26 | - | + |
| 27 | - | - |
| 28 | + | - |
| 29 | - | - |
| 30 | + | - |
| 31 | - | - |
| 32 | - | - |
| 33 | + | - |
| 34 | - | - |
| 35 | - | - |
| 36 | - | - |
| 37 | - | - |
| 38 | - | - |
| 39 | - | - |
| 40 | - | - |
| 41 | - | - |
| 42 | - | + |
| 43 | - | - |
| 44 | - | - |
| 45 | + | - |
| 46 | + | - |
| 47 | + | - |
| 48 | + | - |
| 49 | + | - |
| 50 | + | - |
5. Discussion
Staphylococcus aureus is one of the most common pathogens that have been isolated from patients in hospitals, especially MRSA strains are among important nosocomial infections for humans (1). Having knowledge about the prevalence of MRSA and their virulence factors is useful for treatment and control of community and hospital acquired S. aureus infections. Following the emergence of MRSA strains, molecular studies showed that some of these isolates carry pvl gene (20). The pvl positive strains lead to infections with different clinical appearance even in immunocompromised patients lead to necrotic pneumonia, which its mortality can be as much as 75% (8). Therefore, frequent monitoring of this pathogen, its antibiotic susceptibility and determining their virulence factors is of great importance in control and treatment of infections.
According to the results of this study, only 30% of isolates identified as MRSA were positive for mecA gene. This explains that it maybe mecA positive MRSA isolates have different mechanism for methicillin resistance than mecA. These results are in agreements with other reports such as reported by Bagdonas et al. in Lithuania (23.4%) (21), Turnidge et al. in Australia (24%) (22), Li et al. in China (12%) (23) and also in different parts of Iran, e.g. Fatholahzadeh et al. in Tehran (36%) (24) and by Japoni et al. in Shiraz (43%) (25). However, the frequency of mecA positive isolates in this study is more than the results of Bagdonas et al. Turnidge et al. and Li et al. while is less than the results obtained by Fatholahzadeh et al. (2009) and Japoni et al. This finding is a hopeful result that the frequency of mecA in Ahvaz is yet less than other studied areas of Iran. The occurrence of pvl gene in previous studies has been reported from 2% - 35% (26, 27). In comparison the obtained results for pvl gene in the present study is near to the lower limit obtained from similar studies. No association between mecA and pvl genes was found in our study. It is possible that mecA and pvl genes but not both of them be present on the chromosome of clinical isolates of S. aureus.
The obtained results in the present study are in agreement with the finding of Okon et al. (27) and Terry Alli et al. (9) that reported no pvl positive strain is there among tested MRSA strains. Okon et al. (27) have suggested it is maybe that resistance determinants be carried on other mobile elements, such as plasmids, transposons, and phages; so, their elimination from bacterial cell would result in the absence of mecA gene and consequently no association with pvl gene. Affolabi et al. (28) also reported no significant association between pvl and mecA in S. aureus isolates from Cotonou. However, the pvl gene has been reported in some MRSA in other studies (29, 30).
According to the results of this study, PCR assay for MRSA gene can be useful for definite diagnosis of MRSA strains. Identification of mecA positive strains can be used as a guide for separating infected patients from others in hospital environment in order to prevent gene transfer among clinical strains and also distribution of virulent factors.
Acknowledgements
References
-
1.
David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23(3):616-87. [PubMed ID: 20610826]. https://doi.org/10.1128/CMR.00081-09.
-
2.
Dash N, Panigrahi D, Al Zarouni M, Yassin F, Al-Shamsi M. Incidence of community-acquired methicillin-resistant Staphylococcus aureus carrying Pantone-Valentine leucocidin gene at a referral hospital in United Arab Emirates. APMIS. 2014;122(4):341-6. [PubMed ID: 23919760]. https://doi.org/10.1111/apm.12150.
-
3.
Sanchini A, Del Grosso M, Villa L, Ammendolia MG, Superti F, Monaco M, et al. Typing of Panton-Valentine leukocidin-encoding phages carried by methicillin-susceptible and methicillin-resistant Staphylococcus aureus from Italy. Clin Microbiol Infect. 2014;20(11):O840-6. [PubMed ID: 24835735]. https://doi.org/10.1111/1469-0691.12679.
-
4.
Carleton HA, Diep BA, Charlebois ED, Sensabaugh GF, Perdreau-Remington F. Community-adapted methicillin-resistant Staphylococcus aureus (MRSA): population dynamics of an expanding community reservoir of MRSA. J Infect Dis. 2004;190(10):1730-8. [PubMed ID: 15499526]. https://doi.org/10.1086/425019.
-
5.
Srinivasan A, Seifried S, Zhu L, Srivastava DK, Flynn PM, Shenep JL, et al. Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus infections in children with cancer. Pediatr Blood Cancer. 2009;53(7):1216-20. [PubMed ID: 19731325]. https://doi.org/10.1002/pbc.22254.
-
6.
Khosravi AD, Hoveizavi H, Farshadzadeh Z. The prevalence of genes encoding leukocidins in Staphylococcus aureus strains resistant and sensitive to methicillin isolated from burn patients in Taleghani Hospital, Ahvaz, Iran. Burns. 2012;38(2):247-51. [PubMed ID: 21924558]. https://doi.org/10.1016/j.burns.2011.08.002.
-
7.
Los FC, Randis TM, Aroian RV, Ratner AJ. Role of pore-forming toxins in bacterial infectious diseases. Microbiol Mol Biol Rev. 2013;77(2):173-207. [PubMed ID: 23699254]. https://doi.org/10.1128/MMBR.00052-12.
-
8.
Yanagihara K, Kihara R, Araki N, Morinaga Y, Seki M, Izumikawa K, et al. Efficacy of linezolid against Panton-Valentine leukocidin (PVL)-positive meticillin-resistant Staphylococcus aureus (MRSA) in a mouse model of haematogenous pulmonary infection. Int J Antimicrob Agents. 2009;34(5):477-81. [PubMed ID: 19665359]. https://doi.org/10.1016/j.ijantimicag.2009.06.024.
-
9.
Terry Alli OA, Ogbolu DO, Mustapha JO, Akinbami R, Ajayi AO. The non-association of Panton-Valentine leukocidin and mecA genes in the genome of Staphylococcus aureus from hospitals in South Western Nigeria. Indian J Med Microbiol. 2012;30(2):159-64. [PubMed ID: 22664430]. https://doi.org/10.4103/0255-0857.96675.
-
10.
Kim MH, Lee WI, Kang SY. Detection of methicillin-resistant Staphylococcus aureus in healthcare workers using real-time polymerase chain reaction. Yonsei Med J. 2013;54(5):1282-4. [PubMed ID: 23918582]. https://doi.org/10.3349/ymj.2013.54.5.1282.
-
11.
Ba X, Harrison EM, Edwards GF, Holden MT, Larsen AR, Petersen A, et al. Novel mutations in penicillin-binding protein genes in clinical Staphylococcus aureus isolates that are methicillin resistant on susceptibility testing, but lack the mec gene. J Antimicrob Chemother. 2014;69(3):594-7. [PubMed ID: 24216768]. https://doi.org/10.1093/jac/dkt418.
-
12.
van der Meeren BT, Millard PS, Scacchetti M, Hermans MH, Hilbink M, Concelho TB, et al. Emergence of methicillin resistance and Panton-Valentine leukocidin positivity in hospital- and community-acquired Staphylococcus aureus infections in Beira, Mozambique. Trop Med Int Health. 2014;19(2):169-76. [PubMed ID: 24205917]. https://doi.org/10.1111/tmi.12221.
-
13.
Neela V, Ehsanollah GR, Zamberi S, Van Belkum A, Mariana NS. Prevalence of Panton-Valentine leukocidin genes among carriage and invasive Staphylococcus aureus isolates in Malaysia. Int J Infect Dis. 2009;13(3):e131-2. [PubMed ID: 18955004]. https://doi.org/10.1016/j.ijid.2008.07.009.
-
14.
Chen FJ, Hiramatsu K, Huang IW, Wang CH, Lauderdale TL. Panton-Valentine leukocidin (PVL)-positive methicillin-susceptible and resistant Staphylococcus aureus in Taiwan: identification of oxacillin-susceptible mecA-positive methicillin-resistant S. aureus. Diagn Microbiol Infect Dis. 2009;65(4):351-7. [PubMed ID: 19766426]. https://doi.org/10.1016/j.diagmicrobio.2009.07.024.
-
15.
Tile PM. Bailey & Scott's Diagnostic Microbiology. New York: Mosby; 2013.
-
16.
Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Wayne; 2006.
-
17.
Performance standards for antimicrobial susceptibility testing; Twenty second informational supplement. Wayne; 2012.
-
18.
Sambrookm J, Fritsch EF, Maniatis T. Molecular cloning, A Laboratorry Manual. New York: Cold Spring Harbor Laboratory Press; 1989.
-
19.
McClure JA, Conly JM, Lau V, Elsayed S, Louie T, Hutchins W, et al. Novel multiplex PCR assay for detection of the staphylococcal virulence marker Panton-Valentine leukocidin genes and simultaneous discrimination of methicillin-susceptible from -resistant staphylococci. J Clin Microbiol. 2006;44(3):1141-4. [PubMed ID: 16517915]. https://doi.org/10.1128/JCM.44.3.1141-1144.2006.
-
20.
Shore AC, Tecklenborg SC, Brennan GI, Ehricht R, Monecke S, Coleman DC. Panton-Valentine leukocidin-positive Staphylococcus aureus in Ireland from 2002 to 2011: 21 clones, frequent importation of clones, temporal shifts of predominant methicillin-resistant S. aureus clones, and increasing multiresistance. J Clin Microbiol. 2014;52(3):859-70. [PubMed ID: 24371244]. https://doi.org/10.1128/JCM.02799-13.
-
21.
Bagdonas R, Tamelis A, Rimdeika R, Kiudelis M. Analysis of burn patients and the isolated pathogens. Lithuanian Surg. 2004;2(3):190-3.
-
22.
Turnidge JD, Nimmo GR, Pearson J, Gottlieb T, Collignon PJ, Australian Group on Antimicrobial R. Epidemiology and outcomes for Staphylococcus aureus bacteraemia in Australian hospitals, 2005-06: report from the Australian Group on Antimicrobial Resistance. Commun Dis Intell Q Rep. 2007;31(4):398-403. [PubMed ID: 18268881].
-
23.
Li J, Zeng H, Xu X. [A study of methicillin -- resistant staphylococcus aureus (MRSA) in a burn unit with repetitive -- DNA -- sequence -- based PCR fingerprinting]. Zhonghua Shao Shang Za Zhi. 2001;17(2):88-90. [PubMed ID: 11876917].
-
24.
Fatholahzadeh B, Emaneini M, Aligholi M, Gilbert G, Taherikalani M, Jonaidi N, et al. Molecular characterization of methicillin-resistant Staphylococcus aureus clones from a teaching hospital in Tehran. Jpn J Infect Dis. 2009;62(4):309-11. [PubMed ID: 19628913].
-
25.
Japoni A, Alborzi A, Orafa F, Rasouli M, Farshad S. Distribution patterns of methicillin resistance genes (mecA) in Staphylococcus aureus isolated from clinical specimens. Iran Biomed J. 2004;8(4):173-8.
-
26.
Melles DC, Gorkink RF, Boelens HA, Snijders SV, Peeters JK, Moorhouse MJ, et al. Natural population dynamics and expansion of pathogenic clones of Staphylococcus aureus. J Clin Invest. 2004;114(12):1732-40. [PubMed ID: 15599398]. https://doi.org/10.1172/JCI23083.
-
27.
Okon KO, Basset P, Uba A, Lin J, Oyawoye B, Shittu AO, et al. Cooccurrence of predominant Panton-Valentine leukocidin-positive sequence type (ST) 152 and multidrug-resistant ST 241 Staphylococcus aureus clones in Nigerian hospitals. J Clin Microbiol. 2009;47(9):3000-3. [PubMed ID: 19605575]. https://doi.org/10.1128/JCM.01119-09.
-
28.
Affolabi D, Odoun M, Faïhun F, Damala RL, Ahovegbe L, Prudence Wachinou A. Non-association of the presence of panton-valentine leukocidin gene with antimicrobial-resistance in Staphylococcus aureus isolates in Cotonou, Benin. Int J of Curr Res. 2014;6(1):4617-20.
-
29.
San Juan JA, Cabugao DBG, Dabbay B, Cabrera EC. Occurrence of Methicillin-Resistant Staphylococcus aureus (MRSA) among the Health Workers of Rizal Provincial Hospital and Characterization for the Presence of luks-lukf PVL Gene. Philippine J Sci. 2012;141(2):157-63.
-
30.
Rebollo-Perez J, Ordonez-Tapia C, Herazo-Herazo C, Reyes-Ramos N. Nasal carriage of Panton Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus in healthy preschool children. Rev Salud Publica (Bogota). 2011;13(5):824-32. [PubMed ID: 22634949].