Abstract
Background:
Cervical cancer is regarded as the second highest cause of cancer deaths in Nigeria, with an overall prevalence similar to most developing countries. Screening for cervical cancer is primarily performed using papanicolaou (PAP) staining procedure, in Nigeria.Objectives:
This study aimed to use human papillomavirus (HPV) DNA typing, as a means of ascertaining the presence of high risk HPV in cytology samples, which are positive for the presence of cervical intraepithelial neoplasia (CIN), using the PAP screening procedure.Patients and Methods:
Amplification of DNA was done using polymerase chain reaction. Gene sequencing was carried out to determine the presence of high risk HPV from cervical smears that were positive for abnormal cytology, from a cross-sectional study involving women between the ages of 16 - 65 years, screened for CIN and cervical cancer, in Lokoja, Nigeria.Results:
Result showed a 100% presence of high risk HPV in all the samples with abnormal cytology. The HPV genotype 35 accounted for the highest percentage of the HPVs cases, with a 40% incidence. The HPV genotype 31 accounted for 30% of samples, while HPV genotype 16 and 18 accounted for 20% and 10% of samples, respectively.Conclusions:
The high prevalence of HPV in abnormal cytology underlines to the fact that the presence of HPV is a critical factor in the development of cervical cancer. The use of HPV DNA techniques could actually become an effective and fast means of ascertaining the presence of HPV in abnormal cytology.Keywords
Uterine Cervical Neoplasms DNA DNA Probes HPV Papanicolaou Test Cervical Intraepithelial Neoplasia
1. Background
Cervical cancer is one of the most critical health issues, affecting women in developing countries. It is regarded as the commonest malignancy of the female genital tract in Nigeria (1). There are 36.59 million Nigerians, aged 15 years and older, who are at risk of cervical cancer (2). Cervical cancer is regarded as the second most common cancer amongst women worldwide, and it accounts for 250,000 deaths and 500,000 new patients worldwide, annually (3). Persistent infection by human papillomavirus (HPV) is considered to be the main causative agent of cervical cancer and other anogenital cancers. Transmission of genital HPV infection occurs predominantly through sexual contact, although the infection is mostly self-limiting and clears within 6 months to 2 years, in most cases. Persistent infection by high risk HPV is a critical factor in the development of cervical cancer. Also, factors that contribute to consistent occurrence of HPV infection and development of cervical cancer in women include the number of sexual partners, the age of initiation of sexual activity and the sexual behaviour of the male partner (4).
Cervical cytology screening, utilizing papanicolaou (PAP) staining techniques, remains the most used method for screening for cervical cancer in Nigeria, despite the high rates of false negatives (5). The use of HPV DNA detection techniques offers higher sensitivity and specificity. The severe restriction on the availability of infrastructure, resources and funding, in developing countries, has made it difficult to adopt HPV DNA detection, as a routine cervical cancer prevention strategy (5). Therefore, this research utilizes these molecular techniques in detecting the presence of HPV in abnormal cytology, rendering it as a confirmatory test to cytology.
Over 80% of cervical cancers in sub Saharan Africa are detected in late stages, which is largely due to the lack of information about cervical cancer and prevention services (3). The control of cervical cancer is directly linked to prevention of HPV infection and identification of precancer cases, through screening and subsequent management of identified invasive diseases. These techniques are generally unavailable in Nigeria. In cases, were cervical cancer screening is performed, the PAP test is often the only screening test done. Consequently, the use of HPV DNA typing, which is a major tool used in developed countries, is hardly utilized in Nigeria.
2. Objectives
This study aimed to use HPV DNA typing, as a means of ascertaining the presence of high risk HPV in cytology samples, positive for cervical intraepithelial neoplasia (CIN), using the PAP screening procedure.
3. Patients and Methods
3.1. Description of Study Area
Lokoja is the administrative capital of Kogi state, Nigeria. Lokoja town has a heterogeneous population comprising the Igala, Ebira, Yoruba, Nupe, Okun, Oworo, Bassa-Nge, Kakanda, Egbura and Hausa ethnic groups. Lokoja town is situated at 7˚49’ North and 6˚44’ East. The town is the most centrally located in Nigeria, at the confluence of the rivers Niger and Benue.
3.2. Study Population
The study was carried out among women attending the Obstetrics and Gynecology Clinic of the Federal Medical Centre, Lokoja, Nigeria, aged 16 - 65 years old, who were willing to participate and met the inclusion criteria. A structured close ended questionnaire was administered to these patients, after an informed consent had been obtained, followed by clinical examination.
3.3. Ethical Consideration
Approval for this study was obtained from the Ethical Review Committee of the Federal Medical Centre, Lokoja. The study did not implicate any cost to the subjects and information from the patients and/or parents/tutors were confidential.
3.4. Sample Collection and Preparation
A structured close ended questionnaire was administered to the participating population, using the convenience sampling technique, after due consent had been taken. Cervical smears were collected by a gynecologist after visual inspection. Sample collection was performed as described by Mbamara et al. (6), each of the participants being placed in a dorsal position, with her legs flexed at the hip and knee and abducted. The labia were parted with gloved thumb and index fingers. A Cusco’s bivalve speculum, which was not lubricated, was then passed and fixed to visualize the cervix, under a bright light source. The hook end of the Ayres’ spatula was inserted into the cervix and swept through 360˚ movements, either in a clockwise or counterclockwise direction, to scrape the entire squamocolumnar junction of the transformation zone. The smears collected were immediately fixed to slides, before being transferred to the laboratory for processing. The fixed smears were stained by a cytotechnologist, using the PAP staining procedure and read by a histopathologist, under a light microscope. The slides were reported as normal, inflammatory, abnormal (epithelial lesion) or unsatisfactory. The abnormal ones were classified using the Bethesda 2001 system. The slides with abnormal smear were then tested for the presence of HPV DNA, with several of the slides with normal cytology, which had been previously confirmed for the absence of HPV, utilized for control.
3.5. DNA Extraction Procedure
To start the molecular analysis, the cover slips on the slides were removed, carefully. This was done by soaking the slides in xylene, for 3 days. After the cover slips were removed, the smears were carefully taken from the slides, using sterile scalpel blades, as described by Sharifah et al. (7). A DNA extraction kit (Integrated DNA Technologies, Coralville, IA, USA) was utilized for DNA extraction. The extracted smears were transferred to 1.5 mL tubes for DNA extraction. Buffy coats, as well as proteinase K, were used to lyse the cells of the organism, with the DNA extraction done using chloroform. Equal volumes of 100% ethanol were added to 20 µL of 3M sodium acetate. The mixture was incubated at -200˚C over night. After incubating overnight, the tubes were centrifuged at maximum speeds, for 10 - 30 minutes, in a cold centrifuge, in the same orientation. The ethanol was removed and remaining the DNA pellet. A volume of 400 µL 70% ethanol was added to the tubes containing the DNA pellets and spun at maximum speed, for 5 minute, at 4˚C. The DNA were dried out by leaving the tube open for 3 - 5 minutes. The pellets were resuspended in 20 - 50 µL sterile water. Volumes of 1 µL, 5 µL, and 10 µL were run on an agarose gel, to estimate quantity and integrity. Quantities of 100 - 800 ng were used for polymerase chain reaction (PCR).
3.6. Gel Electrophoresis Procedure
Due to a 1.5% thickness required, 3 g of agarose were used. The agarose was heated in solution in a microwave, until it was completely dissolved. This was then allowed to cool in a water bath, set at 50 - 55˚C. The required numbers of combs were placed in the gel tray. A volume of 5 µL of ethidium bromide was added to the cooled gel and poured into a gel tray. This mixture was allowed to cool for 15 - 30 minutes, at room temperature. The combs were removed and placed in electrophoresis chamber and covered with tris-acetate-EDTA (TAE) buffer. The DNA and standard (ladder) were loaded onto gel and this was electrophoresed for at least 1 hour. The DNA bands were visualized, using a gel imaging system (Gel Doc 2000, Bio-Rad Laboratories Inc., Hercules, CA, USA).
3.7. The Polymerase Chain Reaction and Sequencing Procedure
The PCR components were produced by integrated DNA technologies (Coralville, IA, USA), and the reaction contained primers Gp5: 5’TTTGTTACTGTGGTAGATACTAC-3’and Gp6: 5’GAAAAATAAACTGTAAATCATATTC-3’. The reaction was programmed as follow: initial denaturing step, at 95˚C, for 15 minute, 10 cycles of 30 second at 94˚C, 90 second at 72˚C, followed by 30 cycles of 30 second at 94˚C, 90 second at 63˚C and 90 second at 72˚C, with final extension at 72˚C, for 10 minute. A DNA band of 150 kb was considered a positive result. The method used for the gene sequencing is the dye termination method.
A labeled sterile 0.5 mL tube for each sample was prepared. A fresh stop solution/glycogen mixture was prepared, as follows, per sequencing reaction: 2 µL of 3M sodium acetate, 2 µL of 100 NmM Na2-EDTA and 1 µL of 20 mg/mL of glycogen (provided in the kit). To each of the labeled tubes, 5 µL of the stop solution/glycogen mixture was added. The sequencing reactions were transferred to each of the appropriately labeled tube and mixed thoroughly. A volume of 60 µL cold 95% (v/v) ethanol from a -20˚C freezer was added and mixed thoroughly. This was immediately centrifuged at 14,000 rpm at 4˚C, for 15 minute. The supernatant was carefully removed with a micropipette, leaving behind a visible pellet. The pellet was rinsed with 200 µL 70% (v/v) ethanol, from a -20˚C freezer, and centrifuged at 14,000 rpm at 4˚C, for a minimum of 2 minute. The supernatant was carefully removed, with a micropipette. The sample was resuspended in 40 µL of the sample loading solution (provided in the kit).
4. Results
The study population for this research comprised of 203 women who met the inclusion criteria. The socio-demographic characteristics of the study population are described in Table 1. The mean age of the study population was 40.38 years. Those between the ages of 16 - 25 years accounted for 4.93% of the study population, while those between the ages of 26 - 35 years, while 36 - 45 years, 46 - 55 years and 56 - 65 years accounted for 29.56%, 34.48%, 23.65% and 7.59% of the study population, respectively.
Age Distribution of the Subjects in Relation to the Acquisition of Human Papyloma Virus Infection a
| Age Interval, y | Values b |
|---|---|
| 16 - 25 | 10 (4.93) |
| 26 - 35 | 60 (29.56) |
| 36 - 45 | 70 (34.48) |
| 46 - 55 | 48 (23.65) |
| 56 - 65 | 15 (7.39) |
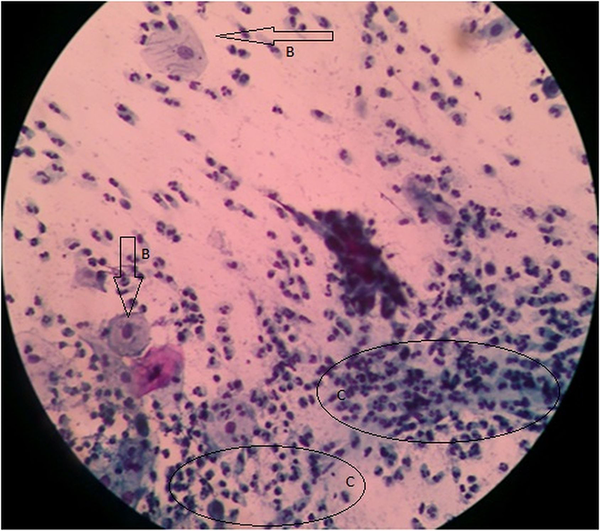
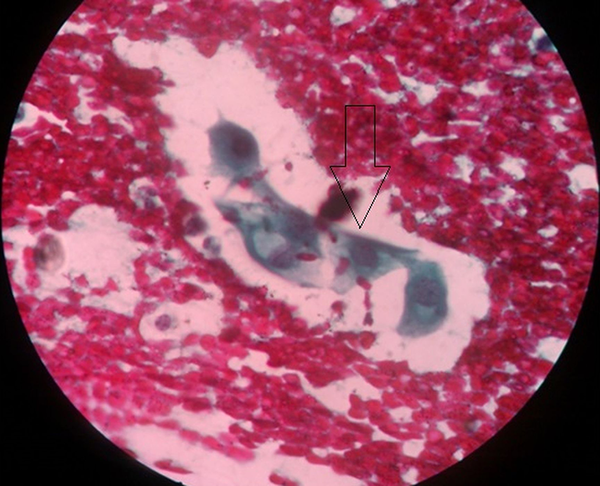
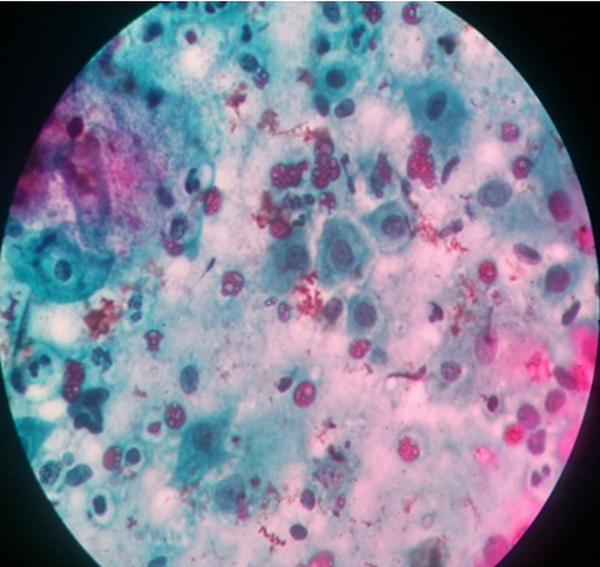
Out of the 203 women, who participated in the study, 193 (95.07%) had normal cytology (they were negative for intraepithelial lesion or malignancy), while 10 (4.93%) women had abnormal cytology. Out of the 10 women with abnormal cytology, four of them were reported as having atypical squamous cells of uncertain significance (ASCUS) (Figure 1), one had squamous cell carcinoma (Figure 2), while the remaining five were reported as having low-grade squamous intraepithelial lesion (Figure 3). Table 2 depicts the prevalence rate of the various CIN found among the respondents. There was no statistical relation between the respondents and type of cervical abnormalities present.
Atypical Squamous Cells of Uncertain Significance of the Cervix Uteri
Squamous Cell Carcinoma of the Cervix Uteri
Low Grade Squamous Intraepithelial Lesion of the Cervix Uteri
| Prevalence | X2 | df | P Value c | |
|---|---|---|---|---|
| Type of Abnormal Cytology | 2.63 | 2 | 0.2685 | |
| ASCUS | 4 (40) | |||
| LSIL | 5 (50) | |||
| SC | 1 (10) |
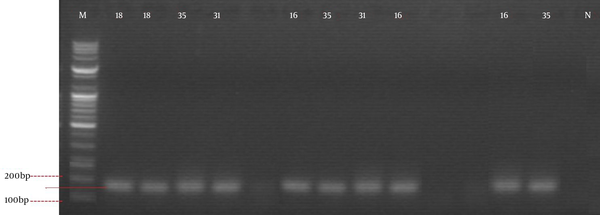
Results also showed that several of the slides had inflammatory smear 16 (7.88%). Two (20%) out of 10 women with abnormal cytology were in the age range of 36 - 45 years, six (60%) of those with abnormal cytology were in the age range of 46 - 55 years, while two (20%) were between 56 - 65 years (Table 1). Sixteen slides were subjected to molecular studies. Two of these slides were those with normal cytology, even though they presented inflammatory cells and the other four had hemorrhage, and these were to serve as experimental controls. After carrying out gene blasting, using the NCBI basic local alignment search tool (BLAST) system, results showed that HPV 35, 31, 18 and 16 strains were present in the sample sequenced, while the samples used as experimental control did not show the presence of HPV DNA (Figure 4).
Positive Polymerase Chain Reaction Bands With Corresponding Human Papyloma Virus Genotypes (1 kb Plus DNA Ladder, 0.9% Agarose Gel Stained With Ethidium Bromide)
5. Discussion
Out of the 203 women who participated in the study, 193 (95.07%) had normal cytology (they were negative for intraepithelial lesion or malignancy), while 10 (4.93%) women had abnormal cytology. The DNA typing was successfully used to confirm the presence of HPV in the abnormal cytology. Genotypic variants of high risk HPV present in this study included HPV genotypes 16, 18, 31, and 35. To a certain extent, this finding is consistent with the type of variants present in West African region, with genotypes 16 and 18, regarded as the most common variants of HPV available worldwide (8). Hamidi-Fard et al. (9) also reported that the most prevalent HPV in their study, in Iran, was HPV 16. Results showed a 100% presence of high risk HPV in all the samples with abnormal cytology, which would have not been known via the conventional PAP test. The HPV genotype 35 had the highest percentage of the high risk HPVs present, with 40% of samples affected. The HPV genotype 31 accounted for 30%, while HPV genotype 16 and 18 accounted for 20% and 10% of samples, respectively. This is similar to the results obtained by Sharifah et al. (7), where 95% of all the abnormal smears, subjected to HPV DNA typing, had high risk HPV present in them.
The cervical smear that had squamous cell carcinoma had HPV genotype 16 present in it. Two of the smears that were presented as atypical squamous cells of undetermined significance (ASCUS) had HPV genotype 35, while the other two smears that had ASCUS were HPV genotypes 31 and 18, respectively. The smears that were presented as low grade squamous intraepithelial lesion (LSIL) had two HPV genotypes 35, two HPV genotypes 31 and one HPV genotype 16, respectively. In similar studies, Sharifah et al. (7) obtained high risk HPV genotypic variants of 16, 18, 31, 51, 52, 56, 58, and 66, from abnormal cervical smears, with the highest frequency of genotypes occurring in LSIL smears. Smits et al. (10) obtained high risk HPV genotypic variants of 16, 18, 31, 33, 45 and 51, from similar abnormal cervical smears. Thomas et al. (11) obtained similar results in Ibadan, Nigeria, where the common high risk HPV genotypic variants found among the subjects were 16, 31, 35, and 58.
The high prevalence of HPV genotypes 31 and 35, in this study, supports suggestions that various genotypic variants are peculiar to particular regions and areas (11). To the best of our knowledge, the only study, concerning genotypic variants of high risk HPV in Nigeria, was conducted in Ibadan, by Thomas et al. (11). They reported that the frequency of HPV types, both in single and multiple infections were combined, and HPV 16 and 35 were the most common high risk types, followed by HPV 31, 58, and 56. However, in our study, which is the first in the North Central Nigeria, the HPV genotype 16 was the variant found in the cervical smear, with squamous cell carcinoma. This has certain significance, as various studies have indicated extensively the role of HPV genotypes 16 and 18 in the development of cervical cancer. The HPV 16 is regarded as a much stronger viral carcinogen, and persists longer than any other type of high risk HPV (8, 12).
The use of DNA typing, as a means of confirming the presence of high risk HPV, is essential to the early detection of cervical cancer, since continuous infection by high risk HPV is a prerequisite for the development of cervical cancer. Early detection is crucial in the fight against this cancer and, therefore, HPV DNA typing provides a fast and effective confirmatory test, compared to the 3 month repetitive test, which is recommended in most cases, and also provides solid information regarding the prevalent HPV genotypes, in a region that is instrumental to the development of vaccines. Adopting molecular techniques, in the routine diagnoses of HPV infection and cervical cancer in Nigeria, is therefore advocated.
Acknowledgements
References
-
1.
Anorlu RI, Igwilo CI, Akanmu AS, Banjo AA, Odunukwe NN, Okany CC, et al. Prevalence of abnormal cervical smears among patients with HIV in Lagos, Nigeria. West Afr J Med. 2007;26(2):143-7. [PubMed ID: 17939318].
-
2.
Adewole IF, Benedet JL, Crain BT, Follen M. Evolving a strategic approach to cervical cancer control in Africa. Gynecol Oncol. 2005;99(3 Suppl 1):S209-12. [PubMed ID: 16202445]. https://doi.org/10.1016/j.ygyno.2005.07.086.
-
3.
World Health Organization. 2012.Prevention of cervical cancer through screening using visual inspection with acetic acid (VIA) and treatment with cryotherapy. A demonstration project in six African countries: Malawi, Madagascar, Nigeria, Uganda, the United Republic of Tanzania, and Zambia. World Health Organization.
-
4.
Kjaer SK, Chackerian B, van den Brule AJ, Svare EI, Paull G, Walbomers JM, et al. High-risk human papillomavirus is sexually transmitted: evidence from a follow-up study of virgins starting sexual activity (intercourse). Cancer Epidemiol Biomarkers Prev. 2001;10(2):101-6. [PubMed ID: 11219765].
-
5.
Fowotade A, Manga MM. Utilization of Human papillomavirus (HPV) DNA detection for cervical cancer screening in developing countries: A myth or reality. Afr J Microbiol Res. 2013;7(20):2135-9.
-
6.
Mbamara SU, Ukah CO, Ikpeze O, Jen O, Onyiaora V. Correlation between visual inspection of the cervix and pap smear test for cervical cancer screening. J Cancer Res Exp Oncol. 2011;3(1):8-13.
-
7.
Sharifah NA, Seeni A, Nurismah MI, Clarence-Ko CH, Hatta AZ, Ho NP, et al. Prevalence of human papillomavirus in abnormal cervical smears in Malaysian patients. Asian Pac J Cancer Prev. 2009;10(2):303-6. [PubMed ID: 19537900].
-
8.
Muñoz N, Franceschi S, Bosetti C, Moreno V, Herrero R, Smith JS, et al. Role of parity and human papillomavirus in cervical cancer: the IARC multicentric case-control study. Lancet. 2002;359(9312):1093-101. https://doi.org/10.1016/s0140-6736(02)08151-5.
-
9.
Hamidi-Fard M, Fattahi-Abdizadeh M, Makvandi M, Ranjbari N, Mansoori E, Samarbaf-Zadeh A. Detection and Genotyping of Human Papillomavirus in Cervical Tissue Samples in Ahvaz, Southwest Iran. Jundishapur J Microbiol. 2013;6(7). eee4569.
-
10.
Smits HL, Tieben LM, Tjong AS, Jebbink MF, Minnaar RP, Jansen CL, et al. Detection and typing of human papillomaviruses present in fixed and stained archival cervical smears by a consensus polymerase chain reaction and direct sequence analysis allow the identification of a broad spectrum of human papillomavirus types. J Gen Virol. 1992;73 ( Pt 12):3263-8. [PubMed ID: 1335027].
-
11.
Thomas JO, Herrero R, Omigbodun AA, Ojemakinde K, Ajayi IO, Fawole A, et al. Prevalence of papillomavirus infection in women in Ibadan, Nigeria: a population-based study. Br J Cancer. 2004;90(3):638-45. [PubMed ID: 14760378]. https://doi.org/10.1038/sj.bjc.6601515.
-
12.
Clifford GM, Gallus S, Herrero R, Munoz N, Snijders PJ, Vaccarella S, et al. Worldwide distribution of human papillomavirus types in cytologically normal women in the International Agency for Research on Cancer HPV prevalence surveys: a pooled analysis. Lancet. 2005;366(9490):991-8. [PubMed ID: 16168781]. https://doi.org/10.1016/S0140-6736(05)67069-9.