Abstract
Background:
Helicobacter pylori infection and related diseases outcome are mediated by a complex interplay between bacterial, host and environmental factors. Several distinct virulence factors of H. pylori have been shown to be associated with different clinical outcomes. Here we focused on vacA and cagA genotypes of H. pylori strains isolated from patients with gastric disorder.Objectives:
The aim of this study was to determine the frequency of two toxins and genotypes of VacA toxin in patients referred to a central hospital in the west of Iran (Imam Reza hospital, Kermanshah) during 2011 - 2012.Patients and Methods:
Samples were collected from patients infected with H. pylori. Gastric biopsy specimens from the stomach antrum and corpus were cultured. PCR analysis was performed for genotyping H. pylorivacA and cagA genes.Results:
Helicobacter pylori was isolated from 48% (96/200) of patients with gastroduodenal disorders. In 81/96 (84%) cases, the cagA gene was present. Among different genotypes of vacA, two s1m2 and s2m2 genotypes were dominant with frequency of 39.5% and 50%, respectively. The frequency of the s1m1 genotype was 7.2% (7/96), which is much lower than elsewhere. H. pylori isolates with positive results for cagA gene and vacA s1m2 genotypes showed statistically significant correlation with peptic ulcer (s1m2 13/34 [38.2%] P = 0.003). However, isolates of H. pylori infection with cagA gene and vacAs2m2 genotypes were significantly associated with development of gastritis (s2m2 41/42 [97.6%] P = 0.000).Conclusions:
About 90% of H. pylori strains potentially contained vacAs2m2 and s1m2 genotypes. Infection with H. pylori strain containing the cagA gene or the vacAs1m1 and s1m2 genotypes was associated with increased incidence of peptic ulcer disease (PUD).Keywords
1. Background
Helicobacter pylori is a Gram-negative comma shaped bacterium, which can cause chronic or acute gastritis, gastric and duodenal ulcers, gastric adenocarcinoma, and mucosa associated lymphoid tissue (MALT) lymphoma. Although most infected individuals may remain asymptomatic (1), H. pylori is colonized in more than 50% of the world’s population (2, 3). The reasons for variation of pathogenesis and prevalence of bacterium might be due to differences in race, health and living standards and more importantly, the virulence factors of the predominant type of the isolated bacteria in the population (4). Among the most important virulence factors we focused on Vacuolating cytotoxin A (VacA) and Cytotoxin-associated gene A (CagA) (5, 6).
VacA is an 88-KD toxin processed from a 140 KD precursor protein (7, 8). The toxin can be found in all H. pylori isolates. The protein constitutes of two variable regions; the signal region (s) located at the N-terminal end of the protein and the middle region (m). There are two major types of s; s1 and s2 and three minor s1 segments; s1a, s1b and s1c. The m region is also known to have two major genotypes: m1 and m2 and two minor subtypes: m1a and m1b. Therefore, the toxin has 4 major and several minor subtypes (9, 10).
CagA is a 125 to 145 kD protein and present in 60 to 90 % of isolates. It is assumed that presence of CagA toxin in a colonized strain implies higher risk of gastritis and gastric cancer (11, 12). In addition to presence and absence of toxin, another important factor in virulence indicator of toxin is structure of protein. The protein consists of several EPIYA motives like EPIYA-ABC or EPIYA-ABD. In western countries with predominant EPIYA-ABC motif isolates, the frequency of gastritis, gastric and duodenal ulcers and gastric cancer is much lower than Far East countries with EPIYA-ABD predominant strains (13). According to published data, the prevalence of H. pylori among Iranians may reach up to 90%, so it is important to predict the outcome of infection within Iranian population (14). As mentioned before, the presence of two toxins and genotypes of VacA is relevant with the appearance of gastric disorders.
2. Objectives
Although there are several reports concerning the prevalence of toxins and genotypes in the Capital and some parts of Iran, we still lack reliable data concerning the aforementioned subjects in the western region of Iran. Therefore, the aim of this study was to determine the frequency of two toxins and genotypes of VacA toxin in patients referred to a central hospital in the west of Iran (Imam Reza hospital, Kermanshah) during 2011 - 2012.
3. Patients and Methods
In this study, 200 patients with gastroduodenal disorders including gastric ulcer, duodenal ulcer and gastritis were enrolled. Every patient with gastroduodenal symptoms referred to endoscopy department of Imam Reza hospital, Kermanshah and patients hospitalized in Internal Medicine (Gastroenterology) were examined by a gastroenterologist. The patients were considered as groups with symptoms of dysphagia, difficulty swallowing, stomach ulcers, reflux, indigestion, and atrophy. The patients who received bismuth drugs, antibiotics and proton pump inhibitors two weeks before sampling and patients with gastrointestinal bleeding were excluded.
3.1. Helicobacter pylori Isolation and DNA Extraction
Biopsy samples were transferred to the laboratory in 1 mL brain heart infusion broth with 25% glycerol within 3 hours. Homogenized samples were cultured on Columbia agar (Merk, Germany) containing 10% egg yolk, 5 mg/L trimethoprim, 10 mg/L vancomycin, 2.5 mg/L amphotericin B and the plates incubated at 37ºC under microaerophilic atmosphere (10% carbon dioxide, 5% oxygen and 85% nitrogen) for 3 to 5 days. Presumptive colonies were confirmed as H. pylori by biochemical tests including catalase, oxidase and urease, and also Gram staining. Confirmed colonies were subcultured on new medium and after incubation the colonies were subjected to DNA extraction by kit according to the manufacturer’s protocol (DNPTM kit, Sinaclon, Iran).
3.2. Polymerase Chain Reaction (PCR)
First H. pylori density was confirmed by PCR analysis using genus and species specific primers targeted 16SrRNA and ureC respectively (Table 1). PCR analyses were performed for determination of vacA s (s1, s2) m (m1, m2), genotypes and also cagA gene using the specific primers (Table 1). cag empty site specific primers were used to confirm the absence of entire cag PAI. PCR reaction was performed in 15 µL volume including 1.5 mM MgCl2, 200 µM of each dNTP, 0.5 µM of each primer, 1X PCR buffer, 1 U Taq DNA polymerase and 100 ng of H. pylori chromosomal DNA. After heating at 95°C for 5 minutes, amplification was performed over 30 cycles of 95°C for 30 seconds, 30 seconds at specific annealing temperature for each primer and 72°C for 30 seconds followed by 72°C for 5 minutes. Annealing temperature were 58°C for gene cagA, 60°C for alleles of vacA and 54°C for cag empty site primers. PCR products were subjected to electrophoresis, stained by ethidium bromide and photography (gel Documentation system, BioRad. Singapore).
List and Characteristic of Oligonucleotide Primers Used in the Study
| Primer Name | Primer Sequence (5’ to 3’) | Product Size, bp | References | |
|---|---|---|---|---|
| 16SrRNA | F | GAT TTT ACC CCT ACA CCA | 422 | (15) |
| R | GCT ATG ACG GGT ATC C | |||
| Hp-ureC | F | CAT CGC CAT CAA AAG CAA AG | 214 | (16) |
| R | CAG AGT TTA AGG ATC GTG TTA G | |||
| cagA | F | GAT AAC AGG CAA GCT TTT GAG G | 349 | (17) |
| R | CTG CAA AAG ATT GTT TGG CAG A | |||
| vacAS1a | F | GTC AGC ATC ACA CCG CAA C | 190 | |
| R | CTG CTT GAA TGC GCC AAA C | |||
| vacAS1b | F | AGC GCC ATA CCG CAA GAG | 187 | |
| R | CTG CTT GAA TGC GCC AAA C | |||
| vacAS2 | F | AGCGCCATACCGCAAGAG | 199 | |
| R | CTG CTT GAA TGC GCC AAA C | |||
| vacAm1 | F | GGT CAA AAT GCG GTC ATG G | 290 | |
| R | CCA TTG GTA CCT GTA GAA AC | |||
| vacAm2 | F | GGA GCC CCA GGA AAC ATT G | 352 | |
| R | CAT AAC TAG CGC CTT GCA C | |||
| vacAm1a | F | GGT CAA AAT GCG GTC ATG G | 290 | |
| R | CTG TTA GTG CCC GCA GAA AC | |||
| vacAm1b | F | GGCCCCAATGCAGTCATGGAT | 291 | |
| R | GCTGTTAGTGCCTAAAGAAGCAT | |||
| cag PAI empty site | F | ACA TTT TGG CTA AAT AAA CGC TG | 550 | (18) |
| R | GGT TGC ACG CAT TTT CCC TTA ATC |
3.3. Statistical Analysis
Statistical analysis of data was performed using logistic regression, chi-square test and Fisher’s exact test, with significance set at a P value of < 0.05. Genotypes with mixed status for vacA were excluded from the calculations of association.
4. Results
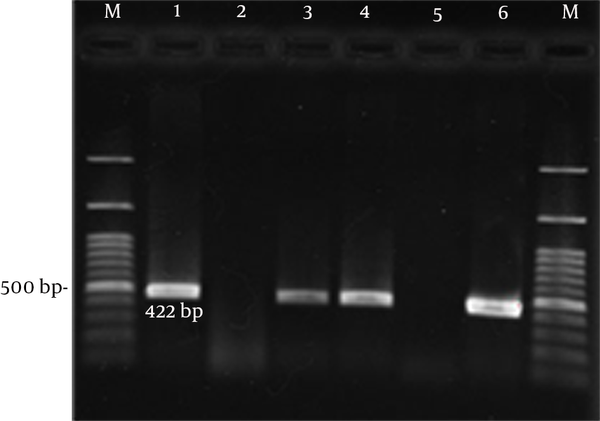
In this study, 200 patients with gastroduodenal difficulties were enrolled; only 96 cases were infected with H. pylori. Age of patients was between 16 and 76 years. Of 96 patients, 41 (42.7%) were female and 55 (47.3%) male. Based on clinical diagnosis, of 96 cases, 20 (20.8%) had peptic ulcer (7 duodenal ulcer and 13 gastric ulcer) and 76 (79.2%) had gastritis. All 96 H. pylori isolates were confirmed by ureC (16) and 16SrRNA PCR (Figure 1).
Gel Electrophoresis of Genus Specific 16SrRNA PCR Products From H. pylori Isolates
4.1. cagA
All genotypes were determined by PCR according to specific primers. Figure 2 shows gel electrophoresis of H. pylori genotyping by cagA, vacA s and m alleles. Overall detection rate of cagA gene in H. pylori isolates was 84.3% (81/96). From 81 cagA positive patients, 45 were male and 36 female. Based on clinical diagnosis, 64 patients were implicated with gastritis, 6 with duodenal ulcers and 11 stomach ulcer. The complete pattern of all positive and negative patients is shown in Table 2.
Gel Electrophoresis of H. pylori Genotyping by cagA, vacA s and m Alleles

Patient Characteristics and Distribution of H. pylorivacA and cagA Genotypes According to the Diseasesa
| Genotypes | Gastritis (n = 76) | Peptic Ulcer (n = 20) | |
|---|---|---|---|
| Gastric ulcer (n = 13) | Duodenal ulcer (n = 7) | ||
| vacAs2 | 48 (63.2) | 1 (7.7) | - |
| vacAs1 | 28 (36.8) | 12 (93.3) | 7 (100) |
| vacAs1a | 26 (34.2) | 12 (92.3) | 7 (100) |
| vacAs1b | 2 (2.6) | - | - |
| vacAm2 | 69 (90.8) | 10 (76.9) | 7 (100) |
| vacAm1 | 7 (9.2) | 3 (23.1) | - |
| vacAm1a | 5 (6.6) | 3 (23.1) | - |
| vacAm1b | 2 (2.6) | - | - |
| cagA | |||
| Positive | 64 (84.2) | 11 (84.61) | 6 (85.75) |
| Negative | 12 (15.8) | 2 (15.49) | 1 (14.3) |
All H. pylori isolates were tested with cag empty site specific primers to confirm the absence of cag PAI. cag empty site PCR positive result found in 21 isolates. Among 21 samples, 6 had positive results for cagA gene and only 15 isolates had true negative findings for cagA gene.
4.2. vacA
vacA gene has variation regions including signal and middle regions classified as s1, s2 and m1, m2 alleles, respectively. However, alleles divided into sub-alleles including s1a, s1b, m1a and m1b. H. Pylori isolates were screened for presence of all sub-alleles by PCR technique. From 96 H. pylori isolates, 47(48.9%) isolates had positive results for s1 allele and 49 (51.1%) for s2. S1a subtype was identified in 45 isolates and s1b subtype in 2 isolates. The presence of m1 was confirmed in 10 (10.5%) isolates, while 86 (89.5%) isolates had positive findings for m2. m1a subtype was identified in 8 isolates and m1b subtype in 2 isolates. Combined forms of suballeles s1m1 genotype were identified in 7.3% of the isolates, s1m2 in 39.5% of the isolates, s2m1 in 3.2% and s2m2 in 50% of the isolates. Presence of cagA gene among s1m1, s1m2, s2m1 and s2m2 positive isolates were 42.8%, 89%, 100%, and 87.5%, respectively.
4.3. Helicobacter pylori Genotypes and Their Association With Gastrointestinal Diseases
Helicobacter pylori isolates with positive results for cagA gene and vacA s1m2 genotypes were significantly correlated with peptic ulcer disease (s1m2 13/34 [38.2%] P = 0.003). However, development of gastritis was significantly associated with H. pylori infection in isolates with cagA gene and vacA s2m2 genotypes (s2m2 41/42 [97.6%] P = 0.000) (Table 3).
Frequency of Helicobacter PylorivacA s1m2 and s2m2 Alleles and cagA Genotypes According to the Diseases
| vacA Alleles | Gastritis (n = 72) | Peptic Ulcer (n = 17) | ||
|---|---|---|---|---|
| cagA+ | cagA- | cagA+ | cagA- | |
| s1m2 | 21 | 1 | 13 | 3 |
| s2m2 | 41 | 6 | 1 | - |
| Total | 62 | 10 | 14 | 3 |
5. Discussion
The rate of infection by H. pylori is averagely 50% in the world population (19). While, the frequency in western countries is approximately 30%, the infection rate in Asia is about 60 to 80% (20). In 0.1 to 1 % of the population, the infection progresses to stomach cancer. In this study, pathogenic properties of H. pylori strains collected from patients with upper gastrointestinal diseases living in the west of Iran were presented. Helicobacter pylori infection is common in Iran, but we lack any information concerning the prevalence of the infection in the west of Iran (21, 22). However, available data suggest that the prevalence is as high as other cities in Iran. The prevalence of vacA genotypes and cagA gene in H. pylori isolates from different parts of the world are different, and there is a direct association between specific genotypes and certain clinical manifestations (23). Our data showed that the prevalence of cagA gene in H. pylori isolates was 84% (81/96). According to published data, the prevalence of cagA in Iranian isolates were 62%, 92% and 68.7% in Tehran, Jahrom and Tabriz, respectively (22, 24, 25). It is obvious that cagA gene frequency is almost the same in different geographic areas in Iran.
Although the prevalence of cagA harboring strains in Iran is similar, the pattern varies among different geographical areas from less than 50% in the central Asia to more than 99% in East Asian countries. This variation could be related to different methodologies like the use of different cagA specific PCR primers and genetic variation of isolates. cagA gene frequency is varied (2, 26, 27). The prevalence of H. pyloricagA positive strains in this study is similar to the reports from Europe and North America with prevalence of 74% and 88%, respectively (28, 29). Many studies reported a higher prevalence of cagA gene in patients with peptic ulcer disease. However, prevalence of cagA positive strains was higher in isolates from patients with peptic ulcer compared with cagA negative strains, even though the difference was not statistically significant.
There are plenty of reports showing a significant association between the prevalence of cagA gene with disease progression; also, the pathogenicity is due to the number of EPIYA motives in CagA protein. The number of EPIYA motives in CagA determines the phosphorylation capability of the protein and higher level of phosphorylation increases inflammatory activity of the toxin. Essentially, all H. pylori strains carry vacA gene, which in certain strains allows in vitro expression of a protein toxin that induces vacuolation of a wide variety of eukaryotic cells. The gene contains both conserved and variable regions, forming a mosaic gene. There are two major regions, called the s (signal sequence-encoding) and m (for mid region-encoding). The VacA toxins with S1 motif are more potent than S2 types (30).
The results of this study showed that the prevalence of s2 and s1 harboring strains are similar. The frequency of s1 positive H. pylori was 40% (19/47) in patients with peptic ulcer disease (PUD) and 60% (28/47) in those with non-ulcer disease (NUD). This is contradictory to other reports which showed that the frequency of this allele in PUD is higher than NUD. For example, in the USA, 90% of patients with duodenal ulcer had s1 allele and the authors concluded that infection with H. pylori strains harboring this allele has an increased risk of developing PUD (31, 32). While, s2 allele frequency was 98% (48/49) in patients with gastritis and only 2% (1/49) in those with PUD.
Among different genotypes of vacA, s1m2 and s2m2 genotypes were dominanted with frequency of (39.5%) s1m2 and (50%) s2m2. In this study, s1m1 genotype frequency was 7.3% (7/96), which is much lower than any other report. The frequency of s1m1 genotype is 24% to 84% worldwide; 26% in Hong Kong, 24% in Nigeria, 48% in Ethiopia, 78% in Korea, 80% in Brazil and 33% in Iran (23, 33-37). There is a direct correlation between toxin activity of different vacA genotypes and pathogenicity of H. pylori. Moreover, the s1m1 genotype has a higher toxicity and can increase gastric atrophy and play an important role in creating gastric ulcer, while it may reduce gastric acid secretion. However, s1m2 and s2m2 genotypes are less potent (38).
The frequency of H. pylori strains harboring s1m1 isolated from patients with gastric ulcer s1m1 genotype was very low in our study. The s2m2 genotypes were more prevalent in our study (50%, 48/96), while it was contrary to other publications from Iran (27%) and also around the world (0% - 57%) (23, 34, 37). So far, the high prevalence of s2m2 has been reported only in North Africa (57%) (30). In this study, s2m2 genotype frequency was 97.9% (47/48) in patients with gastritis and 2.1% (1/48) in those with peptic ulcer. These rates are different with two other published studies reporting 11.9% and 38% (39, 40). Most studies found a significant association between genotype cagA+ and s2m2vacA, but in this study, there was no significant association between cagA+ and s2m2 or cagA- with s2m2. All the s2m1 positive strains had positive results for cagA gene. Moreover, 87.5%, 89% and 42.8% of H. pylori strains with the vacAs2m2, s1m2 and s1m1 genotypes contained the cagA gene as well. Interestingly, among cagA negative strains, three s1m2 harboring strains caused peptic ulcer and three s1m1 harboring strains were related to gastritis.
In conclusion, in patients with gastric disorders in Kermanshah, we found high rates of H. pylori infection, gastritis and PUD. Over 85% of H. pylori organisms carry cagA gene, indicating the presence of putative cag PAI virulence marker. In addition, about 90% of H. pylori strains potentially contained vacAs2m2 and s1m2 genotypes. Infection with H. pylori strain containing the cagA gene or the vacAs1m1 and s1m2 genotypes was associated with increased incidence of PUD. We showed that association between cagA gene and s1m1 genotype was more significant than s1m2 genotype in development of gastric ulcer disease. It seems that s1m1cagA+ strains could predispose development of gastric ulcer in Kermanshah.
Acknowledgements
References
-
1.
Malaty HM, Nyren O. Epidemiology of Helicobacter pylori infection. Helicobacter. 2003;8 Suppl 1:8-12. [PubMed ID: 14617212].
-
2.
Nassrolahei M, Khalilian A. Seropositivity of antibodies against Helicobacter pylori and hepatitis A virus in Iran. Ann Saudi Med. 2004;24(1):61-4. [PubMed ID: 15310023].
-
3.
Suzuki R, Shiota S, Yamaoka Y. Molecular epidemiology, population genetics, and pathogenic role of Helicobacter pylori. Infect Genet Evol. 2012;12(2):203-13. [PubMed ID: 22197766]. https://doi.org/10.1016/j.meegid.2011.12.002.
-
4.
Cave DR. Transmission and epidemiology of Helicobacter pylori. Am J Med. 1996;100(5A):12S-7S. [PubMed ID: 8644777].
-
5.
Arents NL, van Zwet AA, Thijs JC, Kooistra-Smid AM, van Slochteren KR, Degener JE, et al. The importance of vacA, cagA, and iceA genotypes of Helicobacter pylori infection in peptic ulcer disease and gastroesophageal reflux disease. Am J Gastroenterol. 2001;96(9):2603-8. [PubMed ID: 11569682]. https://doi.org/10.1111/j.1572-0241.2001.04104.x.
-
6.
Posselt G, Backert S, Wessler S. The functional interplay of Helicobacter pylori factors with gastric epithelial cells induces a multi-step process in pathogenesis. Cell Commun Signal. 2013;11:77. [PubMed ID: 24099599]. https://doi.org/10.1186/1478-811X-11-77.
-
7.
Cover TL, Blaser MJ. Purification and characterization of the vacuolating toxin from Helicobacter pylori. J Biol Chem. 1992;267(15):10570-5. [PubMed ID: 1587837].
-
8.
Ilver D, Barone S, Mercati D, Lupetti P, Telford JL. Helicobacter pylori toxin VacA is transferred to host cells via a novel contact-dependent mechanism. Cell Microbiol. 2004;6(2):167-74. [PubMed ID: 14706102].
-
9.
Sheu SM, Hung KH, Sheu BS, Yang HB, Wu JJ. Association of nonsynonymous substitutions in the intermediate region of the vacA gene of Helicobacter pylori with gastric diseases in Taiwan. J Clin Microbiol. 2009;47(1):249-51. [PubMed ID: 19020071]. https://doi.org/10.1128/JCM.01651-08.
-
10.
Jones KR, Jang S, Chang JY, Kim J, Chung IS, Olsen CH, et al. Polymorphisms in the intermediate region of VacA impact Helicobacter pylori-induced disease development. J Clin Microbiol. 2011;49(1):101-10. [PubMed ID: 21084502]. https://doi.org/10.1128/JCM.01782-10.
-
11.
Backert S, Tegtmeyer N, Selbach M. The versatility of Helicobacter pylori CagA effector protein functions: The master key hypothesis. Helicobacter. 2010;15(3):163-76. [PubMed ID: 20557357]. https://doi.org/10.1111/j.1523-5378.2010.00759.x.
-
12.
Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4(9):688-94. [PubMed ID: 15343275]. https://doi.org/10.1038/nrc1433.
-
13.
Sahara S, Sugimoto M, Vilaichone RK, Mahachai V, Miyajima H, Furuta T, et al. Role of Helicobacter pylori cagA EPIYA motif and vacA genotypes for the development of gastrointestinal diseases in Southeast Asian countries: a meta-analysis. BMC Infect Dis. 2012;12:223. [PubMed ID: 22994150]. https://doi.org/10.1186/1471-2334-12-223.
-
14.
Alikhani MY, Arebestani MR, Sayedin Khorasani M, Majlesi A, Jaefari M. Evaluation of Helicobacter pylori vacA and cagA Genotypes and Correlation With Clinical Outcome in Patients With Dyspepsia in Hamadan Province, Iran. Iran Red Crescent Med J. 2014;16(11). eee19173. [PubMed ID: 25763216]. https://doi.org/10.5812/ircmj.19173.
-
15.
Pena JA, Fox JG, Ferraro MJ, Versalovic J. Molecular resistance testing of Helicobacter pylori in gastric biopsies. Arch Pathol Lab Med. 2001;125(4):493-7. [PubMed ID: 11260622]. https://doi.org/10.1043/0003-9985(2001)125<0493:MRTOHP>2.0.CO;2.
-
16.
Alvandi A, Abiri R, Aryan E, Rezaei M, Bagherabadi S. High frequency of Helicobacter pylori DNA in drinking water in Kermanshah, Iran, during June–November 2012. J Water Health. 2014;12(3):504-12. [PubMed ID: 25252354].
-
17.
Chen XJ, Yan J, Shen YF. Dominant cagA/vacA genotypes and coinfection frequency of H. pylori in peptic ulcer or chronic gastritis patients in Zhejiang Province and correlations among different genotypes, coinfection and severity of the diseases. Chin Med J (Engl). 2005;118(6):460-7. [PubMed ID: 15788126].
-
18.
Mukhopadhyay AK, Kersulyte D, Jeong JY, Datta S, Ito Y, Chowdhury A, et al. Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India. J Bacteriol. 2000;182(11):3219-27. [PubMed ID: 10809703].
-
19.
Go MF. Review article: natural history and epidemiology of Helicobacter pylori infection. Aliment Pharmacol Ther. 2002;16 Suppl 1:3-15. [PubMed ID: 11849122].
-
20.
Peterson WL, Fendrick AM, Cave DR, Peura DA, Garabedian-Ruffalo SM, Laine L. Helicobacter pylori-related disease: guidelines for testing and treatment. Arch Intern Med. 2000;160(9):1285-91. [PubMed ID: 10809031].
-
21.
Malekzadeh R, Derakhshan MH, Malekzadeh Z. Gastric cancer in Iran: epidemiology and risk factors. Arch Iran Med. 2009;12(6):576-83. [PubMed ID: 19877751].
-
22.
Souod N, Kargar M, Doosti A, Ranjbar R, Sarshar M. Genetic Analysis of cagA and vacA Genes in Helicobacter Pylori Isolates and Their Relationship with Gastroduodenal Diseases in the West of Iran. Iran Red Crescent Med J. 2013;15(5):371-5. [PubMed ID: 24349721]. https://doi.org/10.5812/ircmj.3732.
-
23.
van Doorn LJ, Figueiredo C, Sanna R, Plaisier A, Schneeberger P, de Boer W, et al. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115(1):58-66. [PubMed ID: 9649459].
-
24.
Ghotaslou R, Milani M, Akhi MT, Nahaei MR, Hasani A, Hejazi MS, et al. Diversity of Helicobacter Pylori cagA and vacA Genes and Its Relationship with Clinical Outcomes in Azerbaijan, Iran. Adv Pharm Bull. 2013;3(1):57-62. [PubMed ID: 24312813]. https://doi.org/10.5681/apb.2013.010.
-
25.
Vaziri F, Najar Peerayeh S, Alebouyeh M, Mirzaei T, Yamaoka Y, Molaei M, et al. Diversity of Helicobacter pylori genotypes in Iranian patients with different gastroduodenal disorders. World J Gastroenterol. 2013;19(34):5685-92. [PubMed ID: 24039362]. https://doi.org/10.3748/wjg.v19.i34.5685.
-
26.
Al Qabandi A, Mustafa AS, Siddique I, Khajah AK, Madda JP, Junaid TA. Distribution of vacA and cagA genotypes of Helicobacter pylori in Kuwait. Acta Trop. 2005;93(3):283-8. [PubMed ID: 15715995]. https://doi.org/10.1016/j.actatropica.2005.01.004.
-
27.
Chomvarin C, Namwat W, Chaicumpar K, Mairiang P, Sangchan A, Sripa B, et al. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA and babA2 genotypes in Thai dyspeptic patients. Int J Infect Dis. 2008;12(1):30-6. [PubMed ID: 17548220]. https://doi.org/10.1016/j.ijid.2007.03.012.
-
28.
Van Doorn LJ, Figueiredo C, Megraud F, Pena S, Midolo P, Queiroz DM, et al. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116(4):823-30. [PubMed ID: 10092304].
-
29.
Yamaoka Y, Kodama T, Gutierrez O, Kim JG, Kashima K, Graham DY. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol. 1999;37(7):2274-9. [PubMed ID: 10364597].
-
30.
Atherton JC, Cao P, Peek RM, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270(30):17771-7. [PubMed ID: 7629077].
-
31.
Gunn MC, Stephens JC, Stewart JA, Rathbone BJ, West KP. The significance of cagA and vacA subtypes of Helicobacter pylori in the pathogenesis of inflammation and peptic ulceration. J Clin Pathol. 1998;51(10):761-4. [PubMed ID: 10023339].
-
32.
Mohammadi M, Oghalaie A, Mohajerani N, Massarrat S, Nasiri M, Bennedsen M, et al. Prevalence of Helicobacter pylori vacuolating cytotoxin and its allelic mosaicism as a predictive marker for Iranian dyspeptic patients. Bull Soc Pathol Exot. 2003;96(1):3-5. [PubMed ID: 12784586].
-
33.
Wong BC, Yin Y, Berg DE, Xia HH, Zhang JZ, Wang WH, et al. Distribution of distinct vacA, cagA and iceA alleles in Helicobacter pylori in Hong Kong. Helicobacter. 2001;6(4):317-24. [PubMed ID: 11843964].
-
34.
Kim SY, Woo CW, Lee YM, Son BR, Kim JW, Chae HB, et al. Genotyping CagA, VacA subtype, IceA1, and BabA of Helicobacter pylori isolates from Korean patients, and their association with gastroduodenal diseases. J Korean Med Sci. 2001;16(5):579-84. [PubMed ID: 11641526].
-
35.
Bojary MR, Foroozandeh M, Alvandi AH, Hashemi SM, Masjedian F, Nazifi A. Study of the CagA gene prevalence in Helicobacter Pylori strains isolated from patients with upper gastrointestinal disorders in Iran. Govaresh. 2004;9(3):176-80.
-
36.
Ashour AA, Magalhaes PP, Mendes EN, Collares GB, de Gusmao VR, Queiroz DM, et al. Distribution of vacA genotypes in Helicobacter pylori strains isolated from Brazilian adult patients with gastritis, duodenal ulcer or gastric carcinoma. FEMS Immunol Med Microbiol. 2002;33(3):173-8. [PubMed ID: 12110479].
-
37.
Hussein NR, Mohammadi M, Talebkhan Y, Doraghi M, Letley DP, Muhammad MK, et al. Differences in virulence markers between Helicobacter pylori strains from Iraq and those from Iran: potential importance of regional differences in H. pylori-associated disease. J Clin Microbiol. 2008;46(5):1774-9. [PubMed ID: 18353934]. https://doi.org/10.1128/JCM.01737-07.
-
38.
Wei GC, Chen J, Liu AY, Zhang M, Liu XJ, Liu D, et al. Prevalence of Helicobacter pylori vacA, cagA and iceA genotypes and correlation with clinical outcome. Exp Ther Med. 2012;4(6):1039-44. [PubMed ID: 23226771]. https://doi.org/10.3892/etm.2012.704.
-
39.
Paniagua GL, Monroy E, Rodriguez R, Arroniz S, Rodriguez C, Cortes JL, et al. Frequency of vacA, cagA and babA2 virulence markers in Helicobacter pylori strains isolated from Mexican patients with chronic gastritis. Ann Clin Microbiol Antimicrob. 2009;8:14. [PubMed ID: 19405980]. https://doi.org/10.1186/1476-0711-8-14.
-
40.
Miernyk K, Morris J, Bruden D, McMahon B, Hurlburt D, Sacco F, et al. Characterization of Helicobacter pylori cagA and vacA genotypes among Alaskans and their correlation with clinical disease. J Clin Microbiol. 2011;49(9):3114-21. [PubMed ID: 21752979]. https://doi.org/10.1128/JCM.00469-11.