Abstract
Background:
Cervical cancer is the leading cause of death from cancer in under-developed countries. Human papilloma virus (HPV) 16 and 18 are the most prevalent types associated with carcinogenesis in the cervix. Conventional Polymerase Chain Reaction (PCR), type-specific and consensus primer-based PCR followed by sequencing, Restriction Fragment Length Polymorphism (RFLP) or hybridization by specific probes are common methods for HPV detection and typing. In addition, some researchers have developed a multiplex PCR for simultaneous detection and typing of different HPVs.Objectives:
The aim of the present study was to investigate the prevalence of HPV infection and its types in cervical Squamous Cell Carcinoma (SCC) using the Nested Multiplex PCR (NMPCR) assay.Patients and Methods:
Sixty-six samples with histologically confirmed SCC were evaluated. Total DNA was isolated by phenol–chloroform extraction and ethanol precipitation. Nested multiplex PCR was performed with first-round PCR by GP-E6/E7 consensus primers for amplification of the genomic DNA of all known mucosal HPV genotypes and second-round PCR by type-specific multiplex PCR primer cocktails.Results:
Human papilloma virus infection was detected in 78.8% of samples, with the highest prevalence of HPV 16 (60.6%) while concurrent infections with two types was detected in 10.6%.Conclusions:
The NMPCR assay is more convenient and easy for analysis of results, which is important for fast diagnosis and patient management, in a type-specific manner.Keywords
Human Papillomavirus 16 Multiplex Polymerase Chain Reaction Carcinoma Squamous Cell
1. Background
Cervical cancer is the second most common cancer in females, worldwide (1). The estimated 470000 new cases of infection and about 230000 deaths per year represents a global challenge, especially in under-developed countries, where cervical cancer is the leading cause of death from cancer (2). It is believed that infection with human papillomaviruses (HPVs) is necessary for development of cervical cancer (3). This infection, which is one of the most common Sexually Transmitted Infections (STIs) worldwide (4), not only contributes to the etiology of cervical cancer, especially in the case of high-risk HPV types (HPV 16, 18, 31, 33, etc.), but also has a causative role in anogenital, as well as some head and neck cancers (5). Human papilloma viruses 16 and 18 are the most prevalent types associated with carcinogenesis in the cervix (55% and 16%, respectively) (6).
Infection with high-risk papilloma viruses might be associated with poor prognosis and increased risk of progression to Cervical Intraepithelial Neoplasia (CIN) (7, 8). The malignancies of the cervix originate mostly from the cervical transformation zone, and metastasis can involve surrounding tissues, including vagina, pelvis or nearby lymph nodes (9). The conventional assay for HPV detection is the Polymerase Chain Reaction (PCR), followed by DNA sequencing for determination of individual types. Moreover, type-specific primers allow for detection of each type, separately, yet several amplification processes are required for each sample (10, 11). Alternatively, using consensus primers such as PGMY-SPF10, GP5+-GP6+ and MY09-MY11 for amplification of a broad spectrum of HPV genotypes, followed by sequencing, restriction fragment length polymorphism analysis or hybridization with type-specific probes increase the sensitivity of genotyping (12-15). However, the process is laborious. As an alternative, a method has been described for simultaneous detection and typing of various HPV types by a Nested Multiplex PCR (NMPCR) assay (16).
2. Objectives
In this study, we aimed to investigate the prevalence of HPV infection and its types in cervical Squamous Cell Carcinoma (SCC) using this NMPCR assay.
3. Patients and Methods
3.1. Samples
A total of 66 patients, with histologically-confirmed SCC, who had undergone surgery at Imam Khomeini Hospital, Tehran, Iran, between years 2005 and 2012, were evaluated. Hematoxylin and eosin-stained slides of each paraffin block were reviewed by an expert pathologist. Routine formalin-fixed and paraffin-embedded material blocks were retrieved from the files of the Department of Pathology of Imam Khomeini Hospital.
3.2. DNA Extraction
Eight-micrometer slices of paraffin-embedded blocks were sectioned and placed in individual sterile autoclaved microcentrifuge tubes. To avoid possible contamination, extreme care was taken during tissue sampling and processing with sterile materials. At first, paraffin was removed by xylene and the samples were digested in lysis buffer (50 mM Tris pH 8.5, 1 mM EDTA, 0.5% Tween), containing 50 μL proteinase K (Qiagen, Germany) at 37°C overnight. Total DNA was isolated by phenol–chloroform extraction and ethanol precipitation (16). The DNA pellets were suspended with 20 μL TE buffer (10 mmol/L Tris-HCl, 1 mmol/L EDTA, pH 8.0) and stored at −20°C until use for the PCR.
3.3. Polymerase Chain Reaction for the Human Papilloma Virus
Human papilloma virus detection was performed as described elsewhere (16). In brief, Nested Multiplex PCR (NMPCR) was performed with first-round PCR by GP-E6/E7 consensus primers (GenScript, Germany) (Table 1), for amplification of the genomic DNA of all known mucosal HPV genotypes, and second-round PCR, by type-specific multiplex PCR primer cocktails. Primers for the identification of high-risk genotypes 16, 18, 31, and 33 and low-risk genotypes 6 and 11 were synthesized (GenScript, Germany). The primers were used in two cocktails (Table 2) and the identification of each HPV type was achieved according to the size of the nested PCR product by gel electrophoresis.
Sequences of GP-E6/E7 Consensus Primers
| Primers | Sequence | Amplicon, bp | Position |
|---|---|---|---|
| GP-E6-3F | GGG WGK KAC TGA AAT CGG T | ||
| HPV 6/11 → 580 | HPV 6/11 → 27 - 607 | ||
| GP-E7-5B | CTG AGC TGT CAR NTA ATT GCT CA | ||
| HPV 16 → 609 | HPV 16 → 27 - 636 | ||
| HPV 18 → 639 | HPV 18 → 35 - 674 | ||
| GP-E7-6B | TCC TCT GAG TYG YCT AAT TGC TC | ||
| HPV 31 → 603 | HPV 31 → 31 - 634 | ||
| HPV 33 → 615 | HPV 33 → 32 - 647 |
Sequences of Type-Specific Nested Polymerase Chain Reaction Primers
| HPV Genotype | Amplicon (bp) | Sequence | Position |
|---|---|---|---|
| I | |||
| 16 | 457 | ||
| CAC AGT TAT GCA CAG AGC TGC | 141 - 161 | ||
| CAT ATA TTC ATG CAA TGT AGG TGT A | 597 - 573 | ||
| 18 | 322 | ||
| CAC TTC ACT GCA AGA CAT AGA | 170 - 190 | ||
| GTT GTG AAA TCG TCG TTT TTC A | 491 - 470 | ||
| 31 | 263 | ||
| GAA ATT GCA TGA ACT AAG CTC G | 137 - 158 | ||
| CAC ATA TAC CTT TGT TTG TCA A | 399 - 378 | ||
| II | |||
| 33 | 398 | ||
| ACT ATA CAC AAC ATT GAA CTA | 172 - 192 | ||
| GTT TTT ACA CGT CAC AGT GCA | 569 - 549 | ||
| 6/11 | 334 | ||
| TGC AAG AAT GCA CTG ACC AC | 201 - 220 | ||
| TGC ATG TTG TCC AGC AGT GT | 534 - 515 | ||
Polymerase chain reactions were performed in a final volume of 50 μL. Each PCR mixture contained 10X PCR buffer, 50 mM MgCl2, 100 μM concentration of each deoxynucleoside triphosphate (dNTP), 5 U/μL of Taq DNA polymerase (Qiagen, Germany), 10 μmol of each primer and pure H2O. Two microliters of the PCR product served as the template for the nested PCRs. Ten microliters of the amplification products were analyzed by electrophoresis on 2% agarose gels and ethidium bromide staining. As an internal control, amplification of a 110-bp product of the human beta-globin housekeeping gene was performed by PCO3 and PCO4 primers (GenScript, Germany). Amplifications were performed with the following cycling profile: a four minutes pre-denaturation step at 94°C, 40 cycles of one minute denaturation at 94°C, one minute annealing at 55°C, and one minute elongation at 72°C. The last cycle was followed by a final extension step of 10 minutes at 72°C.
The conditions for the PCRs with E6 consensus primers were 94°C for one minute, 40°C for one minute, and 72°C for two minutes for a total of 40 amplification cycles. The first cycle was preceded by a four minutes denaturation step at 94°C. The last cycle was followed by an additional 10-minute elongation step at 72°C. Nested Multiplex PCRs with type-specific primers were performed under the following conditions: 35 cycles of 94°C for 30 seconds, 56°C for 30 seconds, and 72°C for 45 seconds. The first cycle was preceded by a four minutes denaturation step and the last cycle was followed by a four minutes elongation step.
3.4. Statistical Analysis
The SPSS software, version 20.0 (SPSS Inc., Chicago) was used for the calculations.
4. Results
4.1. Polymerase Chain Reaction for the Human Papilloma Virus
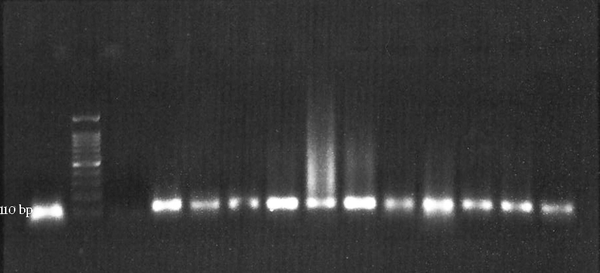
Polymerase chain reaction results for beta-globin housekeeping gene were positive for all samples (Figure 1).
Beta-Globin Gene Amplification
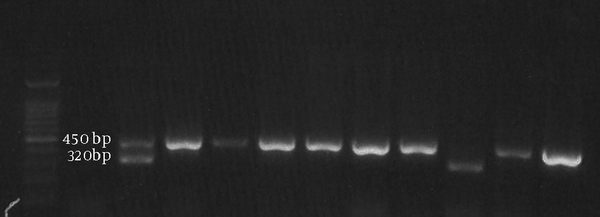
Human papilloma virus detection was performed by NMPCR (Figure 2) and the results are summarized in Figure 3.
Agarose Gel Electrophoresis for Nested Multiplex Polymerase Chain Reaction Products of Cocktail-I
Human Papilloma Virus Typing by the Nested Multiplex Polymerase Chain Reaction Assay
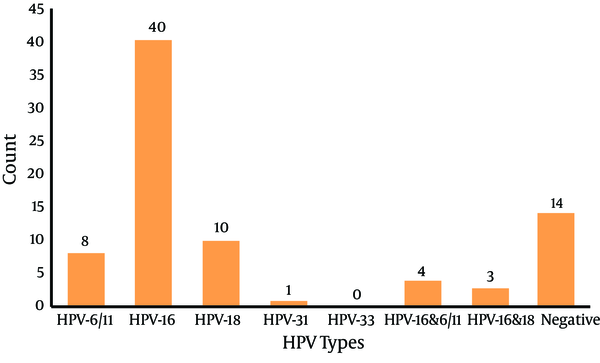
In this study, 78.8% (52/66) of samples were positive for HPV infection, with HPV 16 having the highest prevalence (40/66) (60.6%) and HPV 31 having the lowest prevalence (1.5%). Human papilloma virus-33 was not detected in these samples. Concurrent infections with two types were detected in 10.6% of samples, where three cases of co-infection with types 16 and 18 and four cases of co-infection with types 16 and 6/11 were detected in our study.
5. Discussion
Over 130 HPV types have been detected, and about 40 HPV types have been isolated from the genital tract, until now (17). Although the outcome of the infection depends on several criteria, including age, persistency, concurrent infections, host genetics, and smoking, the most effective factor could possibly be the type of HPV. An infection with high-risk HPV types is essential for development of cancer (18). The most common assay for HPV detection is based on the PCR procedure and subsequent typing. Typing is achieved by different methods such as, hybridization by specific probes, restriction fragment length polymorphisms and direct DNA sequencing (19).
In order to accomplish a more convenient assay, a type-specific multiplex PCR was performed in order to detect four high-risk and two low-risk HPV types. In this way, detection and typing of HPV infection are achieved simultaneously. Although, the HPV genome has been detected in 99.7% of cervical cancers (20, 21), in this study, 78.8% of samples were HPV infected. This deviation is probably due to incomplete coverage of all high-risk and low-risk HPV types in our assay. On the other hand, 69.7% of samples were positive for high-risk types 16 and 18, which is in complete consistency with about 71% prevalence of these two types in cervical carcinoma samples (22). Of all cervical SCC samples, 90.4% were infected with high-risk genotypes, while 9.6% were positive for low-risk HPVs (ratio, 9 to 1). These results are consistent with other reports, regarding the prevalence of high-risk types of HPV in Iran (23-25). In addition, concurrent infection with two HPV types was detected in 10.6% of samples that may result in more severe outcome of the infection.
In conclusion, type-specific multiplex PCR for simultaneous detection and typing of HPV infection may result in more convenient and easy analysis of the results, which is important for fast diagnosis and patient management, in a type-specific manner.
Acknowledgements
References
-
1.
Insinga RP, Liaw KL, Johnson LG, Madeleine MM. A systematic review of the prevalence and attribution of human papillomavirus types among cervical, vaginal, and vulvar precancers and cancers in the United States. Cancer Epidemiol Biomarkers Prev. 2008;17(7):1611-22. [PubMed ID: 18628412]. https://doi.org/10.1158/1055-9965.EPI-07-2922.
-
2.
Reshmi G, Pillai MR. Beyond HPV: Oncomirs as new players in cervical cancer. FEBS Lett. 2008;582(30):4113-6. [PubMed ID: 19032954]. https://doi.org/10.1016/j.febslet.2008.11.011.
-
3.
Fabiano V, Mariani L, Giovagnoli MR, Raffa S, Vincenzoni C, de Michetti F, et al. Cervical cancer screening program based on HPV testing and conventional Papanicolaou cytology for jail inmates. Health. 2010;2(9):1027-32. https://doi.org/10.4236/health.2010.29151.
-
4.
Smith JS, Melendy A, Rana RK, Pimenta JM. Age-specific prevalence of infection with human papillomavirus in females: A global review. J Adolesc Health. 2008;43(4 Suppl):S5-25. [PubMed ID: 18809145]. https://doi.org/10.1016/j.jadohealth.2008.07.009.
-
5.
Lepique AP, Rabachini T, Villa LL. HPV vaccination: The beginning of the end of cervical cancer? - A Review. Mem Inst Oswaldo Cruz. 2009;104(1):1-10. [PubMed ID: 19274369].
-
6.
Castellsague X. Natural history and epidemiology of HPV infection and cervical cancer. Gynecol Oncol. 2008;110(3 Suppl 2):S4-7. [PubMed ID: 18760711]. https://doi.org/10.1016/j.ygyno.2008.07.045.
-
7.
Cox JT, Lorincz AT, Schiffman MH, Sherman ME, Cullen A, Kurman RJ. Human papillomavirus testing by hybrid capture appears to be useful in triaging women with a cytologic diagnosis of atypical squamous cells of undetermined significance. Am J Obstet Gynecol. 1995;172(3):946-54. [PubMed ID: 7892889].
-
8.
Cuzick J, Szarewski A, Terry G, Ho L, Hanby A, Maddox P, et al. Human papillomavirus testing in primary cervical screening. Lancet. 1995;345(8964):1533-6. [PubMed ID: 7791438].
-
9.
Sarraf Z, Hamedi B, Hooshmand S, Mosalaie A, Robati M, Momtahan M, et al. The effect of extrafascial hysterectomy after completion of external beam radiotherapy for treatment of locally advanced stages (IIB-III) of Cervical Cancer. Iran Red Crescent Med J. 2013;15(12). eeee10758. [PubMed ID: 24693381]. https://doi.org/10.5812/ircmj.10758.
-
10.
Baay MF, Quint WG, Koudstaal J, Hollema H, Duk JM, Burger MP, et al. Comprehensive study of several general and type-specific primer pairs for detection of human papillomavirus DNA by PCR in paraffin-embedded cervical carcinomas. J Clin Microbiol. 1996;34(3):745-7. [PubMed ID: 8904451].
-
11.
van den Brule AJ, Claas EC, du Maine M, Melchers WJ, Helmerhorst T, Quint WG, et al. Use of anticontamination primers in the polymerase chain reaction for the detection of human papilloma virus genotypes in cervical scrapes and biopsies. J Med Virol. 1989;29(1):20-7. [PubMed ID: 2555442].
-
12.
Gravitt PE, Peyton CL, Apple RJ, Wheeler CM. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J Clin Microbiol. 1998;36(10):3020-7. [PubMed ID: 9738060].
-
13.
Hildesheim A, Schiffman MH, Gravitt PE, Glass AG, Greer CE, Zhang T, et al. Persistence of type-specific human papillomavirus infection among cytologically normal women. J Infect Dis. 1994;169(2):235-40. [PubMed ID: 8106758].
-
14.
Jacobs MV, Snijders PJ, van den Brule AJ, Helmerhorst TJ, Meijer CJ, Walboomers JM. A general primer GP5+/GP6(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J Clin Microbiol. 1997;35(3):791-5. [PubMed ID: 9041439].
-
15.
Sasagawa T, Minemoto Y, Basha W, Yamazaki H, Nakamura M, Yoshimoto H, et al. A new PCR-based assay amplifies the E6-E7 genes of most mucosal human papillomaviruses (HPV). Virus Res. 2000;67(2):127-39. [PubMed ID: 10867192].
-
16.
Sotlar K, Diemer D, Dethleffs A, Hack Y, Stubner A, Vollmer N, et al. Detection and typing of human papillomavirus by e6 nested multiplex PCR. J Clin Microbiol. 2004;42(7):3176-84. [PubMed ID: 15243079]. https://doi.org/10.1128/JCM.42.7.3176-3184.2004.
-
17.
Munoz N, Castellsague X, de Gonzalez AB, Gissmann L. Chapter 1: HPV in the etiology of human cancer. Vaccine. 2006;24 Suppl 3:S3/1-10. [PubMed ID: 16949995]. https://doi.org/10.1016/j.vaccine.2006.05.115.
-
18.
Schiffman M, Castle PE, Jeronimo J, Rodriguez AC, Wacholder S. Human papillomavirus and cervical cancer. Lancet. 2007;370(9590):890-907. [PubMed ID: 17826171]. https://doi.org/10.1016/S0140-6736(07)61416-0.
-
19.
Romero-Pastrana F. Detection and typing of human papilloma virus by multiplex PCR with type-specific primers. ISRN Microbiol. 2012;2012:186915. [PubMed ID: 23724318]. https://doi.org/10.5402/2012/186915.
-
20.
Bosch FX, Manos MM, Munoz N, Sherman M, Jansen AM, Peto J, et al. Prevalence of human papillomavirus in cervical cancer: A worldwide perspective. International biological study on cervical cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995;87(11):796-802. [PubMed ID: 7791229].
-
21.
Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189(1):12-9. [PubMed ID: 10451482]. https://doi.org/10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F.
-
22.
World Health Organisation. Human Papillomavirus and Related Diseases Report. Barcelona: WHO/ICO Information Centre on HPV and Cervical Cancer; Available from: http://www.HPVcentre.net/statistics/reports/XWX.pdf.
-
23.
Khorasanizadeh F, Hassanloo J, Khaksar N, Mohammad Taheri S, Marzaban M, Rashidi BH, et al. Epidemiology of cervical cancer and human papilloma virus infection among Iranian women - analyses of national data and systematic review of the literature. Gynecol Oncol. 2013;128(2):277-81. [PubMed ID: 23200918]. https://doi.org/10.1016/j.ygyno.2012.11.032.
-
24.
Shahsiah R, Khademalhosseini M, Mehrdad N, Ramezani F, Nadji SA. Human papillomavirus genotypes in Iranian patients with cervical cancer. Pathol Res Pract. 2011;207(12):754-7. [PubMed ID: 22041132]. https://doi.org/10.1016/j.prp.2011.09.011.
-
25.
Jalilvand S, Shoja Z, Nourijelyani K, Tohidi HR, Hamkar R. Meta-analysis of type-specific human papillomavirus prevalence in Iranian women with normal cytology, precancerous cervical lesions and invasive cervical cancer: Implications for screening and vaccination. J Med Virol. 2015;87(2):287-95. [PubMed ID: 25156655]. https://doi.org/10.1002/jmv.24053.