Abstract
Background:
On March 31, 2013, human infections with a novel avian influenza A (H7N9) virus were firstly reported in China. By February 7, 2015, a total of 562 laboratory-confirmed human cases of H7N9 infection were reported in mainland China.Objectives:
This study aimed to determine whether there is a novel H7N9 virus infection in human and animals in Taizhou city, China, in 2013.Methods:
In this study, we developed real-time reverse transcription-polymerase chain reaction (RT-PCR) assays to detect H7N9 virus infection in human and animals (chicken, pig, and pigeon) in Taizhou city in the beginning of H7N9 epidemics of 2013. Also, we studied the novel isolated H7N9 virus strain by using genome sequencing and phylogenetic analysis.Results:
Of 150 samples tested, 7 were detected to be H7N9 positive which all were collected from chickens. Also, a novel H7N9 virus strain (A/chicken/jiangsu/1021/2013) was isolated and studied. Phylogenetic analysis showed the novel isolated H7N9 virus had high identity to the four novel human H7N9 viruses (A/Shanghai/1/2013, A/Shanghai/2/2013, A/Anhui/1/2013, and A/Hangzhou/1/2013).Conclusions:
Our results demonstrated a novel H7N9 virus in chickens in Taizhou city in 2013 that may pose a potential risk to public health.Keywords
Influenza A Virus H7N9 Subtype Real-Time Polymerase Chain Reaction Evolution Molecular China
1. Background
Influenza A virus belongs to the family Orthomyxoviridae with an eight-segmented, single-stranded, negative-sense RNA genome (1, 2). Based on differences within antigenic properties of the two surface glycoproteins, i.e. hemagglutinin (HA) and neuraminidase (NA), 18 HA (H1 - H18) and 11 NA (N1 - N11) subtypes have been identified (3-5). Avian influenza virus (AIV) is a kind of influenza A viruses (such as H5N1, H7N1, and H9N2) which can affect several species of domestic poultry, such as chickens, turkeys, quails, and ducks, as well as wild birds (6-8). Based on the severity of diseases in poultry, AIV can generally be divided into two groups: low pathogenic (LPAI) that typically causes subclinical infections and highly pathogenic (HPAI) that can cause severe infections with high mortality rates in poultry (9, 10).
On March 31, 2013, the Chinese public health authorities notified the world health organization (WHO) of 3 cases (2 cases in Shanghai, 1 case in Anhui) of human infections with novel avian influenza A (H7N9) virus (11, 12). On 4 April 2013, the Chinese ministry of agriculture found the similar H7N9 virus infection in pigeons and chickens in the wet markets of Shanghai (13). The emergence of novel H7N9 virus epidemics causes a significant threat to the public health. By February 7, 2015, a total of 562 confirmed human H7N9 infection cases were reported in mainland China (14).
In this study, we developed real-time RT-PCR assays using two pairs of primers recommended by the WHO to detect H7N9 virus infection in human and animals (chicken, pig, and pigeon) in Taizhou city in 2013. Taizhou city is in the centre of the Yangtze River Delta Region, which was the epicenter of the 2013 H7N9 epidemic (Figure 1). Of 150 samples tested, 7 were detected to be positive using the H7N9 real-time RT-PCR assays. Furthermore, we isolated a novel H7N9 virus (A/chicken/jiangsu/1021/2013) from chicken samples collected in a live poultry market, and studied its genetic characteristics using genome sequencing and phylogenetic analysis.
2. Objectives
In this research, samples collected from human and animals were tested. Also, we isolated a novel H7N9 virus (A/chicken/jiangsu/1021/2013) and studied its genetic characteristics using genome sequencing and phylogenetic analysis.
3. Methods
3.1. Ethics Statement
All research involving human participants was approved by the institutional review boards of the people’s hospital of Taizhou and Taizhou institute of virology (No.: TZ2013003), in accordance with the guidelines for the protection of human subjects. Written informed consent was obtained from each participant.
3.2. Biosafety Procedures
Laboratory procedures for the propagation of H7N9 virus were performed in the Laboratory P3 at the Wuhan University with the required containments and safety precautions. The samples treatment and RNA isolation were performed in the biosafety 2 laboratory at the Taizhou institute of virology.
3.3. Samples Collection and RNA Isolation
A total of 150 samples were collected in Taizhou city from April 8 to May 4, 2013. We collected 31 clinical of samples throat swabs from patients with influenza like illness (ILI) or pneumonias by medical practitioners of the people’s hospital of Taizhou. We also collected throat swabs from chickens (58 cases) in market and pigs (51 cases) in farm and message pigeons (10 cases). Throat swabs were suspended in 3 mL Dulbecco’s modified Eagle medium (DMEM, Invitrogen, CA, USA). Total RNAs were isolated from 140 µL samples using the QIAamp viral RNA mini extraction kit (Qiagen, Shanghai, China) in accordance with the manufacturer’s protocol, and final 60 µL RNA was stored at -80°C until use.
3.4. Real-Time RT-PCR Assay
The primers used in the assays for targeting the HA and NA genes of H7N9 were those recommended by WHO (www.who.int/influenza/gisrs_laboratory/cnic_realtime_rt_pcr_protocol_a_h7n9.pdf). The real-time RT-PCR was performed in Bio-Rad iCycler iQ5™ real time PCR system (Bio-Rad, Hercules, CA, USA) using Takara One Step PrimeScript™ RT-PCR Kit (DRR064A, Takara, Dalian, China). Each reaction (25 µL) comprised 5 μL total RNAs, 0.5 μL Ex Taq HS, 0.5 μL PrimeScript RT Enzyme Mix II,12.5 μL 2 × One Step RT-PCR Buffer; the final reaction mixture contained 400 nmol/L of each primer and 200 nmol/L of the probe. The protocol for real-time RT-PCR was as follows: 45°C for 5 minutes, 95°C for 10 seconds, and 45 cycles at 95°C for 5 seconds and 55°C for 45 seconds. The fluorescence was read at the end of 55°C step, which allowed a continuous monitoring of the amount of RNA. All the reactions were performed in duplicate.
3.5. Virus Isolation and Identification
Viruses were isolated by culturing samples that were detected positive by H7N9 real-time RT-PCR assays in 9-day-old embryonated SPF chicken eggs. The eggs were incubated at 37°C for 2 - 3 days, and the allantoic fluids from embryos were then collected and tested for hemagglutinin (HA) activity. The isolates were subtyped by hemagglutination inhibition (HI) and neuraminidase inhibition (NI) assays. All experiments with the infectious, highly pathogenic viruses were conducted under BSL-3 containment.
3.6. RT-PCR Amplification and Genomic Sequencing
The RNA extraction from allantoic fluid was carried out according to the trizol reagent manufacturer’s instructions (Invitrogen, CA, USA). The eight segments of genome sequences were amplified by conventional PCR with specific primer sets described previously (15). The amplified PCR products were separated on 1.5% agarose gels and stained with Gelred and methyl blue. The PCR products were purified and subsequently subjected to bi-directional DNA sequencing in Sangon, Inc. (Sangon, Shanghai, China). The GenBank accession numbers of the HA and NA sequences of the H7N9 virus strain reported in this study are KF938945 and KF938946. Nucleotide BLASTn analysis (http://www.ncbi.nlm.nih.gov/BLAST) was used to identify related reference viruses.
3.7. Phylogenetic Analyses
To understand the evolutionary characterization of H7N9 viruses isolated in this study, the genome sequences of different strains of influenza viruses were obtained from the EpiFlu database of the Global initiative on sharing all influenza data (GISAID) and the GenBank database. Pair-wise sequence alignments were performed with the Megalign program (DNASTAR, Wisconsin, USA) to determine nucleotide sequence similarities. Phylogenetic analyses of the aligned sequences for HA and NA genes were performed by the neighbor-joining method with 1000 bootstraps using MEGA version 5.2 (16).
4. Results
4.1. Design of the Study
Taizhou city is in the center of the Yangtze River Delta Region which was the epicenter of the 2013 H7N9 epidemic (Figure 1). Therefore, it is important to know whether there is novel H7N9 virus infection in human and animals in this area. Thus, we collected 150 samples from human (31 cases), chickens (58 cases), pigs (51 cases), and message pigeons (10 cases) in Taizhou city between April 8 and May 4, 2013. Based on the gained information and collected samples, two different sets of analyses were carried out. First, we carried out real-time RT-PCR to analyze the samples for the presence of the sequences of H7N9 virus. Second, we performed virus isolation and conventional PCR to study its genetic characterization using genome sequencing and phylogenetic analysis.
Geographic Location of Taizhou City, Jiangsu Province, China
4.2. Prevalence of the Novel H7N9 Virus in Chicken Samples Collected in Taizhou City
Two sets of primers, which specifically amplified the sequences of HA-7 and NA-9, were used to detect the presence of novel H7N9 virus. A panel of 150 samples was detected by H7N9 real-time RT-PCR assays which were carried out in duplicate for each sample and repeated three times. Of the 150 samples, 7 were positive for the presence of novel H7N9 influenza virus in the H7N9 real-time RT-PCR assays (Table 1). Further isolation of the novel H7N9 virus from these positive samples confirmed the presence of the H7N9 virus.
Results of the Novel H7N9 Virus Detection in 150 Samples by Real-Time RT-PCR Assays
| Samples | Total | Positive | Negative |
|---|---|---|---|
| Human | 31 | 0 | 31 |
| Chicken | 58 | 7 | 51 |
| Pig | 51 | 0 | 51 |
| Pigeon | 10 | 0 | 10 |
| Total | 150 | 7 | 143 |
4.3. Virus Isolation and Identification
Seven viruses’ strains with HA activity were isolated from samples which were positive in real-time RT-PCR assays. The HI and NI assays indicated that they belonged to the H7N9 subtype, which were also confirmed by genomic sequencing and BLASTn analysis. One virus strain (A/chicken/jiangsu/1021/2013) isolated from a sample of chicken with no clinical symptoms collected in live poultry market of Taizhou city on April 2013 was selected to represent all seven viruses and then subjected to further genetic analysis.
4.4. Phylogenetic Analysis of the Isolated H7N9 Virus Strain
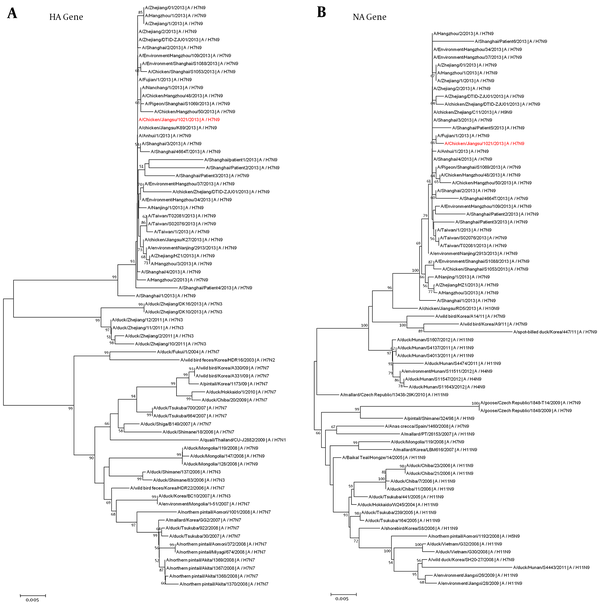
To investigate the genotype and genetic origin of the isolated H7N9 virus (A/chicken/jiangsu/1021/2013), the genome sequences were sequenced. The nucleotide sequences obtained in this study have been deposited in the GenBank database (accession numbers KF938945 and KF938946). The genetic origin of the novel isolated H7N9 virus was initially studied by BLAST analysis. The results showed that the isolated H7N9 virus shares the highest sequence homology with other H7N9 viruses in human and chickens circulating in China since 2013 (> 99.8%) (Table 2). Furthermore, phylogenetic trees were constructed by using MEGA software with the neighbor joining algorithm to study the genetic origin of the isolated virus. The HA and NA sequences of influenza virus reference strains were downloaded from the EpiFlu database and GenBank database. The phylogenetic analysis revealed that the novel isolated H7N9 virus strain (A/chicken/jiangsu/1021/2013) is grouped into the same cluster of H7N9 subtype and also it has a close evolutionary relationship with H7N9 lineage that had been commonly found in China during the H7N9 outbreak in 2013 (Figure 2A and B). These results indicate that the novel H7N9 virus circulated in chicken in Taizhou city in 2013.
Nucleotide Identity of Novel Influenza A (H7N9) Virus Genes and Reference Strains Available in GenBank
| Gene | Virus With Greatest Similarity | GenBank Accession No. | Similarity (%) |
|---|---|---|---|
| PB2 | A/chicken/Zhejiang/DTID-ZJU01/2013(H7N9) | KC899666 | 99.9 |
| PB1 | A/Shanghai/5190T/2013(H7N9) | KF997837 | 99.8 |
| PA | A/chicken/Jiangsu/S002/2013(H7N9) | CY146915 | 99.9 |
| HA | A/Fujian/1/2013(H7N9) | KC994453 | 100 |
| NP | A/Shanghai/02/2013(H7N9) | KF021598 | 99.9 |
| NA | A/Fujian/1/2013(H7N9) | KC994454 | 99.8 |
| M | A/Nanjing/1/2013(H7N9) | KC896777 | 99.8 |
| NS | A/Fujian/1/2013(H7N9) | KC994456 | 99.8 |
Phylogenetic Trees for the HA (A) and NA (B) Genes of the Novel Isolated Virus (A/Chicken/Jiangsu/1021/2013) and Related Reference Influenza Viruses
4.5. Molecular Characterization of H7N9 Virus Isolated From Chicken in Taizhou City
Based on the deduced amino acid sequence, the isolated H7N9 virus strain (A/chicken/jiangsu/1021/2013) contained the multibasic amino acid motif PEIPKGR/GL at their HA cleavage sites, suggesting that this virus strain is LPAI. The amino acid sequence of the receptor-binding site (RBS, residues 105-319) in the HA gene segment of the human or avian A (H7N9) virus determines the preference for human (SA-α-2, 6-Gal) or avian (SA-α-2, 3-Gal) type receptors and by interacting with the receptor, it plays a curial role in the binding of influenza virus to the target cells (13, 17, 18). In this study, the isolated H7N9 virus contained G186V and Q226L mutations at RBS site which could increase avian H5 and H7 viruses’ binding ability to the human type receptors (18-21). (A/Shanghai/2/2013) and (A/Anhui/1/2013) encode the same mutations, whereas (A/Hangzhou/1/2013) encodes G186V and Q226I, which is also found in seasonal influenza A (H3N2) viruses (Table 3).
Amino Acid Comparison of PB2, HA, NA, and NS1 of the Novel Isolated H7N9 Virus and Reference Viruses
| Virus | PB2 | HA | NA | NS1 | |||
|---|---|---|---|---|---|---|---|
| 627 | 186 | 226 | 69 - 73 | 294 | 42 | 218 - 230 | |
| A/chicken/jiangsu/1021/2013 (H7N9) | E | V | L | Deletion | R | L | Deletion |
| Shanghai/1/2013(H7N9) | K | G | Q | Deletion | K | S | Deletion |
| Shanghai/2/2013(H7N9) | K | V | L | Deletion | R | S | Deletion |
| Anhui/1/2013(H7N9) | K | V | L | Deletion | R | S | Deletion |
| Hangzhou/1/2013(H7N9) | K | V | I | Deletion | R | S | Deletion |
| Human influenza virus | K | G | I | No deletion | R | S | No deletion |
| Avian influenza virus | E | G | Q | No deletion | R | S/A | No deletiona |
Neuraminidase (NA) is a glycoside hydrolase enzyme, acting at the final stage of infection. It cleavages sialic acid from cell surface and propagates virions to facilitate the release of virions from infected cells. The length of its stalk is associated with the virulence. It had been reported that a deletion of five amino acids (residues 69 - 73) in its stalk may be associated with its increased virulence in mammals (22, 23). All four human H7N9 virus strains and the novel isolate H7N9 virus have this deletion, suggesting that the novel isolated H7N9 virus may have the same virulence in mammals as four human H7N9 viruses. At the site 294 of the NA gene, (A/Shanghai/1/2013) it has the R294K mutation, which is known to confer resistance to NA inhibitors in N2 and N9 subtype viruses (24-26). But the novel isolated H7N9 virus and the other three human H7N9 viruses did not have this mutation (Table 3).
The mutant E627K of the polymerase PB2 protein is essential for the efficient replication of avian influenza viruses in mammals (11, 27-29). After analyzing amino acid sequences, we found that the four human H7N9 viruses had this mutation but the novel isolated H7N9 virus did not (Table 3).
The NS1 is a multifunctional protein that functions in several ways to defeat the cellular innate immune response in the viral life cycle. Both the novel isolated H7N9 virus and the four human H7N9 viruses could encode a deletion at positions 218 - 230 (the C-terminal PDZ domain-binding motif). The lack of this motif may attenuate these viruses in mammals (30, 31). All four human H7N9 viruses encode NS1 - 42S. In contrast, the novel isolated H7N9 virus encodes NS1 - 42L (Table 3) with the mutation S42L in NS1 protein; however, its significance for the biological properties is currently unclear. Therefore, we will focus on this mutation in our future studies.
5. Discussion
Between 31 March 2013 and February 7, 2015, a total of 562 confirmed cases of human infection with avian influenza A (H7N9) virus was reported in mainland China (14). Human H7N9 infection cases have been reported from Anhui, Jiangsu, Zhejiang, Henan, Fujian, Hunan, Jiangxi, Hebei, Guangdong, Shandong, Guangxi, Guizhou, and the municipalities of Beijing and Shanghai; most of affected areas were the cities and provinces around the Yangtze River Delta Region where the first case of infection was identified (11, 12). In the previous studies, sporadic human infections by other subtypes of H7 virus (e.g. H7N2, H7N3, and H7N7) have been reported in Europe and North America (32). This is the first time that human infection with the avian influenza A (H7N9) subtype is reported. We found that various characteristics of this novel H7N9 virus are different from those of previously reported.
The infection with H7N9 virus can result in severe and fatal respiratory disease which signifies the threat for public health. Therefore, it is great urgent to develop rapid and specific methods for early detection of the novel H7N9 virus infection (33, 34). In this study, we developed real-time RT-PCR assays using two pairs of primers recommended by WHO for detecting H7N9 virus infection in human and animals (chicken, pig, and pigeon) in Taizhou city in 2013. Of 150 samples tested, 7 were detected to be H7N9 positive. Also, we isolated a novel H7N9 virus (A/chicken/jiangsu/1021/2013) from chicken of a poultry market in Taizhou city, and studied its genetic characteristics using genome sequencing and phylogenetic analysis.
Taizhou city is located in the central Jiangsu province which is the geographical centre of the Yangtze River Delta Region (Figure 1). By February 7, 2015, 70 cases of human infection with avian influenza A (H7N9) were reported in Jiangsu province, accounting for about 12.4% (70/562) of all cases in China. Although, there was no official report of human infection of H7N9 cases in Taizhou city in the 2013 (two H7N9 Human infection cases reported in the Spring, 2014), it is important to know whether there is novel H7N9 virus infection in human and animals in Taizhou city. Thus, we collected 150 samples from human (31 cases), chickens (58 cases), pigs (51 cases), and message pigeons (10 cases) in Taizhou city between April 8 and May 4, 2013.
All of samples were detected by real-time RT-PCR assay. Our results showed that 7 samples (7/150, 4.7%) all collected from chickens in the market were detected to be H7N9 positive in real-time RT-PCR assays. H7N9 viruses were also isolated from these positive samples that confirmed the presence of the H7N9 virus circulated in the chicken in the market. We also observed that there are no obvious clinical signs in the chicken seller and people around the market. The reason why chicken vendors who were exposed frequently to H7N9 virus-infected chicken remained infection-free is unclear (34, 35). There was no detection of H7N9 infection in human, pigs, and message pigeons in our study. We will continue our surveillance of H7N9 infection in human and animals in the future studies.
Infectious diseases are the results of interaction of virus, host, and environment. Host factors can include age, sexuality, and underlying medical disorders, while environmental factors comprise temperature, humidity, and poultry exposure status (34). The molecular mechanism of virulence and host adaptation of the novel H7N9 virus remains unclear and may involve viral genes and host factors. Previous studies have shown that the pathogenicity of influenza virus depends on the functional integrity of each gene that is optimal for infection (30, 31, 36-38). For example, hemagglutinin (HA) plays an important role in determining the host species tropism.
In our study, the isolated H7N9 virus contained G186V and Q226L mutations at the RBS site, which could increase its binding ability to the human type receptors (20, 39). Neuraminidase (NA) is crucial for virus release and replication. Studies indicated that a deletion of five amino acids (residues 69 - 73) in NA stalk may be associated with its increased virulence in mammals (22, 23). The isolated H7N9 virus had a same deletion in the stalk region of NA, which may explain an adaptation of H7N9 viruses from aquatic birds to terrestrial poultry such as chickens. The mutant E627K of the polymerase PB2 protein is essential for the efficient replication of avian influenza viruses in mammals. After analyzing amino acid sequences, we found that the isolated H7N9 virus is free of this mutation. This maybe a reason why people in contact with novel H7N9 virus infected chicken did not get infected.
In conclusion, before our study, no cases of H7N9 infection in human or animals in the city of Taizhou have been previously reported (two H7N9 Human infection cases reported in the spring 2014). Our results in this study provided the first direct evidence toward a novel H7N9 virus circulating in chicken in Taizhou city in 2013. Phylogenetic analyses suggest that our isolated novel H7N9 virus strain (A/chicken/jiangsu/1021/2013) is closely related to those currently circulating in human in mainland China. Further study on their ability to replicate as well as their pathogenesis will facilitate the control and prevention of infections due to novel H7N9 virus.
Acknowledgements
References
-
1.
Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56(1):152-79. [PubMed ID: 1579108].
-
2.
Rezaei F, Mirshafiey A, Shahmahmoodi S, Shoja Z, Ghavami N, Mokhtari-Azad T. Influenza Virus-like Particle Containing Two Different Subtypes of Hemagglutinin Confers Protection in Mice Against Lethal Challenge With A/PR8 (H1N1) and A/HK (H3N2) Viruses. Iran Red Crescent Med J. 2013;15(1):75-82. [PubMed ID: 23487492]. https://doi.org/10.5812/ircmj.6252.
-
3.
Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, et al. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol. 2005;79(5):2814-22. [PubMed ID: 15709000]. https://doi.org/10.1128/JVI.79.5.2814-2822.2005.
-
4.
Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, et al. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A. 2012;109(11):4269-74. [PubMed ID: 22371588]. https://doi.org/10.1073/pnas.1116200109.
-
5.
Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, et al. New world bats harbor diverse influenza A viruses. PLoS Pathog. 2013;9(10):1003657. [PubMed ID: 24130481]. https://doi.org/10.1371/journal.ppat.1003657.
-
6.
Alexander DJ. A review of avian influenza in different bird species. Vet Microbiol. 2000;74(1-2):3-13. [PubMed ID: 10799774]. https://doi.org/10.1016/S0378-1135(00)00160-7.
-
7.
Dugan VG, Chen R, Spiro DJ, Sengamalay N, Zaborsky J, Ghedin E, et al. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008;4(5):1000076. [PubMed ID: 18516303]. https://doi.org/10.1371/journal.ppat.1000076.
-
8.
Suarez DL. Evolution of avian influenza viruses. Vet Microbiol. 2000;74(1-2):15-27. [PubMed ID: 10799775]. https://doi.org/10.1016/S0378-1135(00)00161-9.
-
9.
Mo IP, Brugh M, Fletcher OJ, Rowland GN, Swayne DE. Comparative pathology of chickens experimentally inoculated with avian influenza viruses of low and high pathogenicity. Avian Dis. 1997;41(1):125-36. [PubMed ID: 9087329]. https://doi.org/10.2307/1592452.
-
10.
Perdue ML, Garcia M, Senne D, Fraire M. Virulence-associated sequence duplication at the hemagglutinin cleavage site of avian influenza viruses. Virus Res. 1997;49(2):173-86. [PubMed ID: 9213392]. https://doi.org/10.1016/S0168-1702(97)01468-8.
-
11.
Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013;368(20):1888-97. [PubMed ID: 23577628]. https://doi.org/10.1056/NEJMoa1304459.
-
12.
Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, et al. Epidemiology of human infections with avian influenza A(H7N9) virus in China. N Engl J Med. 2014;370(6):520-32. [PubMed ID: 23614499]. https://doi.org/10.1056/NEJMoa1304617.
-
13.
Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, et al. Genetic analysis of novel avian A(H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill. 2013;18(15):20453. [PubMed ID: 23594575].
-
14.
Liu W, Yang K, Qi X, Xu K, Ji H, Ai J, et al. Spatial and temporal analysis of human infection with avian influenza A(H7N9) virus in China, 2013. Euro Surveill. 2013;18(47). [PubMed ID: 24300887].
-
15.
Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146(12):2275-89. [PubMed ID: 11811679]. https://doi.org/10.1007/s007050170002.
-
16.
Kumar S, Tamura K, Nei M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput Appl Biosci. 1994;10(2):189-91. [PubMed ID: 8019868]. https://doi.org/10.1093/bioinformatics/10.2.189.
-
17.
Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440(7083):435-6. [PubMed ID: 16554799]. https://doi.org/10.1038/440435a.
-
18.
Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog. 2010;6(9):1001081. [PubMed ID: 20824086]. https://doi.org/10.1371/journal.ppat.1001081.
-
19.
Zhou J, Wang D, Gao R, Zhao B, Song J, Qi X, et al. Biological features of novel avian influenza A (H7N9) virus. Nature. 2013;499(7459):500-3. [PubMed ID: 23823727]. https://doi.org/10.1038/nature12379.
-
20.
Ramos I, Krammer F, Hai R, Aguilera D, Bernal-Rubio D, Steel J, et al. H7N9 influenza viruses interact preferentially with alpha2,3-linked sialic acids and bind weakly to alpha2,6-linked sialic acids. J Gen Virol. 2013;94(Pt 11):2417-23. [PubMed ID: 23950563]. https://doi.org/10.1099/vir.0.056184-0.
-
21.
Srinivasan K, Raman R, Jayaraman A, Viswanathan K, Sasisekharan R. Quantitative description of glycan-receptor binding of influenza A virus H7 hemagglutinin. PLoS One. 2013;8(2):49597. [PubMed ID: 23437033]. https://doi.org/10.1371/journal.pone.0049597.
-
22.
Sun Y, Tan Y, Wei K, Sun H, Shi Y, Pu J, et al. Amino acid 316 of hemagglutinin and the neuraminidase stalk length influence virulence of H9N2 influenza virus in chickens and mice. J Virol. 2013;87(5):2963-8. [PubMed ID: 23269805]. https://doi.org/10.1128/JVI.02688-12.
-
23.
Matsuoka Y, Swayne DE, Thomas C, Rameix-Welti MA, Naffakh N, Warnes C, et al. Neuraminidase stalk length and additional glycosylation of the hemagglutinin influence the virulence of influenza H5N1 viruses for mice. J Virol. 2009;83(9):4704-8. [PubMed ID: 19225004]. https://doi.org/10.1128/JVI.01987-08.
-
24.
McKimm-Breschkin JL, Sahasrabudhe A, Blick TJ, McDonald M, Colman PM, Hart GJ, et al. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors. J Virol. 1998;72(3):2456-62. [PubMed ID: 9499107].
-
25.
Sleeman K, Guo Z, Barnes J, Shaw M, Stevens J, Gubareva LV. R292K substitution and drug susceptibility of influenza A(H7N9) viruses. Emerg Infect Dis. 2013;19(9):1521-4. [PubMed ID: 23965618]. https://doi.org/10.3201/eid1909.130724.
-
26.
McKimm-Breschkin J, Trivedi T, Hampson A, Hay A, Klimov A, Tashiro M, et al. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob Agents Chemother. 2003;47(7):2264-72. [PubMed ID: 12821478]. https://doi.org/10.1128/AAC.47.7.2264-2272.2003.
-
27.
Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293(5536):1840-2. [PubMed ID: 11546875]. https://doi.org/10.1126/science.1062882.
-
28.
Mok CK, Lee HH, Lestra M, Nicholls JM, Chan MC, Sia SF, et al. Amino acid substitutions in polymerase basic protein 2 gene contribute to the pathogenicity of the novel A/H7N9 influenza virus in mammalian hosts. J Virol. 2014;88(6):3568-76. [PubMed ID: 24403592]. https://doi.org/10.1128/JVI.02740-13.
-
29.
Zhang H, Li X, Guo J, Li L, Chang C, Li Y, et al. The PB2 E627K mutation contributes to the high polymerase activity and enhanced replication of H7N9 influenza virus. J Gen Virol. 2014;95(Pt 4):779-86. [PubMed ID: 24394699]. https://doi.org/10.1099/vir.0.061721-0.
-
30.
Jackson D, Hossain MJ, Hickman D, Perez DR, Lamb RA. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci U S A. 2008;105(11):4381-6. [PubMed ID: 18334632]. https://doi.org/10.1073/pnas.0800482105.
-
31.
Jiao P, Tian G, Li Y, Deng G, Jiang Y, Liu C, et al. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol. 2008;82(3):1146-54. [PubMed ID: 18032512]. https://doi.org/10.1128/JVI.01698-07.
-
32.
Belser JA, Bridges CB, Katz JM, Tumpey TM. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg Infect Dis. 2009;15(6):859-65. [PubMed ID: 19523282]. https://doi.org/10.3201/eid1506.090072.
-
33.
Gao HN, Lu HZ, Cao B, Du B, Shang H, Gan JH, et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013;368(24):2277-85. [PubMed ID: 23697469]. https://doi.org/10.1056/NEJMoa1305584.
-
34.
Skowronski DM, Janjua NZ, Kwindt TL, De Serres G. Virus-host interactions and the unusual age and sex distribution of human cases of influenza A(H7N9) in China, April 2013. Euro Surveill. 2013;18(17):20465. [PubMed ID: 23647627].
-
35.
Han J, Jin M, Zhang P, Liu J, Wang L, Wen D, et al. Epidemiological link between exposure to poultry and all influenza A(H7N9) confirmed cases in Huzhou city, China, March to May 2013. Euro Surveill. 2013;18(20). [PubMed ID: 23725866].
-
36.
Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci U S A. 2005;102(51):18590-5. [PubMed ID: 16339318]. https://doi.org/10.1073/pnas.0507415102.
-
37.
Qi X, Pan Y, Qin Y, Zu R, Tang F, Zhou M, et al. Molecular characterization of avian-like H1N1 swine influenza a viruses isolated in Eastern China, 2011. Virol Sin. 2012;27(5):292-8. [PubMed ID: 23055004]. https://doi.org/10.1007/s12250-012-3262-9.
-
38.
Neumann G, Macken CA, Kawaoka Y. Identification of amino acid changes that may have been critical for the genesis of A(H7N9) influenza viruses. J Virol. 2014;88(9):4877-96. [PubMed ID: 24522919]. https://doi.org/10.1128/JVI.00107-14.
-
39.
Peiris JS, de Jong MD, Guan Y. Avian influenza virus (H5N1): a threat to human health. Clin Microbiol Rev. 2007;20(2):243-67. [PubMed ID: 17428885]. https://doi.org/10.1128/CMR.00037-06.