Abstract
Background:
Keeping in mind the commercial application of polygalacturonase (PG) in juice and beverages industry, bacterial strains were isolated from rotten fruits and vegetables to screen for competent producers of PG.Objectives:
In this study, the plate method was used for preliminary screening of polygalacturonase-producing bacteria, while the Dinitrosalicylic Acid (DNS) method was used for quantifications of PG.Materials and Methods:
Biochemically-identified polygalacturonase-producing Bacillus and Pseudomonas species were further characterized by molecular markers. The genetic diversity among these selected strains was analyzed by investigating microsatellite distribution in their genome. Out of 110 strains, 17 competent strains of Bacillus and eight strains of Pseudomonas were selected, identified and confirmed biochemically. Selected strains were characterized by 16S rRNA sequencing and data was submitted to the national center for biotechnology information (NCBI) website for accession numbers.Results:
Among the Bacillus, Bacillusvallismortis (JQ990307) isolated from mango was the most competent producer of PG; producing up to 4.4 U/µL. Amongst Pseudomonas, Pseudomonasaeruginosa (JQ990314) isolated from oranges was the most competent PG producer equivalent to B. vallismortis (JQ990307). To determine genetic diversity of different strains of Pseudomonas and Bacillus varying in PG production, fingerprinting was done on the basis of Simple Sequence Repeats (SSR) or microsatellites. The data was analyzed and a phylogenetic tree was constructed using the Minitab 3 software for comparison of bacterial isolates producing different concentrations of PG. Fingerprinting showed that presence or absence of certain microsatellites correlated with the ability of PG production.Conclusions:
Bacteria from biological waste were competent producers of PG and must be used on an industrial scale to cope with the demand of PG in the food industry.Keywords
Polygalacturonase Phylogenetic DNA Dinitrosalicylic Acid (DNS) Method Bacillus spp. Pseudomonas spp.
1. Background
Microbial enzymes have had tremendous industrial applications for decades. Pectic substances form the major components of middle lamella and primary plant cell wall. Pectic substances consist of protopectins, pectinic acids, pectins and pectic acids. The main chain of pectin is a partially methyl esterified 1, 4-D-galacturonan. Demethylated pectin is known as pectic acid (pectate) or polygalacturonic acid. Pectin serves as line of defense against pathogens. Phytopathogens contain large quantities of pectinase, especially Polygalacturonase (PG), which facilitates invasion in plant tissue by degrading pectin. These phytopathogens are mostly fungus and are competent producers of PG and help in plant biomass recycling (1). Pectinases are being used largely in industrial processes, especially in the food industry and textile industry, and their demand is being constantly increased (2). Pectinases are a heterogeneous group of enzymes that contribute to 10% of the overall enzyme production and almost 25% of food enzyme throughout the world (3).
Fungal enzymes are not stable at high temperatures, considering that intracellular harvesting is difficult and pasteurization is required to inhibit the growth of mesophilic contaminants (4). Due to the relatively low temperature stability of the fungal enzyme, maceration is carried out at temperatures below 45°C. To inhibit the growth of mesophilic microbes, pasteurization is an essential step of maceration (4). Fungus PG showed best activity at acidic pH and bacterial PG is required when enzyme activity required at neutral pH (5). Keeping in mind the easy harvesting, low cost, high production and activity of PG at wide pH ranges, environmental bacteria are commonly screened for PG.
2. Objectives
The current study aimed to isolate potent PG-producing strains of Bacillus and Pseudomonas, isolated from plant-based waste material and to correlate PG production with molecular markers.
3. Materials and Methods
3.1. Isolation and Purification of Bacterial Isolates
The surface of rotten fruits and vegetables was washed thoroughly with tap water, sterilized by using 70% ethanol. One-gram samples of different fruits and vegetables were pooled and homogenized in sterile water to prepare a 1% final concentration. Three serial dilutions (10-1, 10-2 and 10-3) of pulp were made in autoclaved water and 50 µL of each dilution was spread on plates containing N-broth media and MacConkey agar. After incubation at 37°C for 24 hours, colonies of different morphological characteristics were purified by repeated streaking. Pure cultures were maintained for enzyme studies and identification as 30% glycerol stock at -20°C.
3.2. Preliminary Screening and Identification of Pectinase-Producing Bacterial Strains
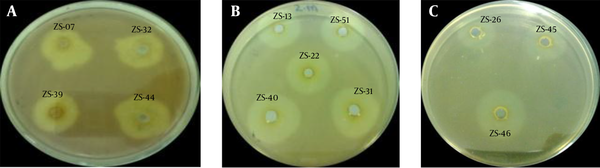
Qualitative determination of pectinase activity was done by inoculating the N-broth medium with bacterial strains followed by incubation at 37°C in a shaking incubator (150 rpm). After 24 hours of incubation, 100 µL of each starter culture with adjusted optical density of 0.1 at 600 nm was inoculated in 5 mL of N-broth media in all test tubes and incubated under the same conditions as above. One milliliter of the overnight culture was centrifuged at 12,000 rpm for 15 minutes to separate the cell mass and the supernatant. Plates containing minimal salt medium supplemented with 0.2% pectin were prepared (6). Wells were made in each petri plate under aseptic conditions as shown in Figure 1 ; 50 µL of the supernatant was added to each well and allowed to diffuse in the medium. The plates were incubated at 37°C for 18 - 24 hours. After incubation, iodine solution (7) was flooded over the plates to detect the zone of inhibition. Morphological and biochemical characterization of the selected bacterial strains was based on the potential of pectinolytic activity.
Pectinase Activity of Various Bacterial Strains
3.3. Enzyme Assay
Polygalacturonase activity of selected bacteria was determined by the colorimetric method using polygalacturonic acid as a substrate of PG and 3-5-dinitrosalicylic acid (DNS) to quantify liberated reducing sugars (8). The reaction mixture containing 1 mL of culture filtrate, 1 mL of 0.9% polygalacturonic acid in 0.1 M of sodium acetate buffer (pH 4.5) was incubated at 45°C. After 30 minutes of incubation, 1.5 mL of DNS solution was added and the mixture was boiled at 100°C for ten minutes. Optical density at 530 nm was measured after cooling the mixture. Controls were run with culture medium. Finally, concentrations of enzyme units were quantified by comparing the optical density with the standard curve of galacturonic acid.
3.4. DNA Extraction and Amplification of 16S rRNA
Genomic DNA was extracted from competent strains of Bacillus and Pseudomonas by the Cetyltrimethylammonium Bromide (CTAB) method (9). Amplification of 16 S rRNA gene of competent strains was done by following the procedure reported by Muyzer et al. (10). Amplified products were sent to the centre for advance molecular biology (CAMB), University of the Punjab, Lahore for sequencing. The obtained sequenced data was blast on national center for biotechnology information (NCBI) and submitted to Bankit for Accession numbers.
3.5. Genetic Diversity of Selected Pectinolytic Bacterial Strains
Genetic diversity of PG-producing strains was determined by DNA finger printing. Six simple sequence repeats (SSR), (GACA)4, (GTG)5, (GATA)4, (CA)8, (GGAT)4 and (CAA)5, were used for PCR amplification. Annealing temperature of all the primers was determined using a gradient PCR reaction. Annealing temperature was 42°C, 48°C and 42°C for (GACA)4, (GTG)5 and (GATA)4, respectively. The remaining primers failed to amplify the target DNA. Polymerase Chain Reaction amplification was performed in a 50 µL reaction mixture containing genomic DNA (100 ng), 25 pmol SSR primer (10 µL), 10X PCR buffer (5 µL), 25 mM MgCl2 (3 µL), 250 mM dNTPs mixture (4 µL) and 5 units of Taq DNA polymerase (0.5 µL) in a total volume of 50 µL.
The amplification was programmed in a thermocycler (Bioer xp cycler, China) as follows: preheating for one minute at 95°C (initial denaturation), 35 cycles each for 20 seconds at 95°C (denaturation), 25 seconds at the annealing temperature (calculated according to the melting temperature (Tm) of each primer), and 20 seconds at 72°C (extension) and a final extension at 72°C for two minutes. Amplification products were separated by gel electrophoresis and photographed fingerprints were transferred to a binary character matrix (one for presence and zero for absence of a band at a particular position). Molecular weights were calculated by comparing the motifs with a standard molecular weight marker (Thermo scientific Cat. # SM 0311, USA). A dendrogram based on the matrix was constructed by cluster analysis with the minitab3 software.
3.6. Statistical Analysis
Each experiment was performed twice, consisting of three replicates. The data was subjected to the Analysis of Variance (ANOVA) test. The means of treatments were grouped by 0.05 probability level on the basis of Duncan’s multiple range test. Graphs were prepared using the Excel Spread Sheet and dendrograms were constructed using Minitab® 3.
4. Results
A total of 110 bacterial strains were isolated from different rotten fruit and vegetable samples.
4.1. Preliminary Screening of Pectinase-Producing bacTeria
Qualitative assay for pectinase activity by the plate method (11) was adopted from preliminary screening of a total of 110 bacterial strains isolated from different fruit and vegetable samples. In the plate method, pectin was present in the culture media as a sole source of carbohydrate and the pectinase activity of the culture medium was determined by the zone of inhibition around the wells. Diameters of the zones were measures in millimeters. Out of 110 strains, 54 were selected for further studies on the basis of pectinase production. The negative control showed no zone of inhibition, which indicated the validity of the results. Competent bacterial strains were identified using culture characteristics, staining reactions and biochemical tests (Catalase, Coagulase, Oxidase, API 20E etc.), following Clinical and Laboratory Standards Institute (CLSI) (2008) guidelines (12). Only Bacillus and Pseudomonas were selected for further studies. For further studies, a total of 22 strains including 17 Bacillus and eight Pseudomonas strains were selected out of 54.
4.2. Quantitative detection of PG in Selected Bacterial Strains
The production of PG by the selective strains of Bacillus and Pseudomonas was quantitatively determined by the DNS method and enzyme activity for each strain of Bacillus and Pseudomonas was calculated by using the standard curve of galacturonic acid. One unit of activity was defined as the amount that released 1 µM of galacturonic acid per minute under standard assay conditions. Some strains of Bacillus and Pseudomonas produced ≥ 4.0 U/µL, which were considered excellent producers of PG whereas others showed variable levels of activity.
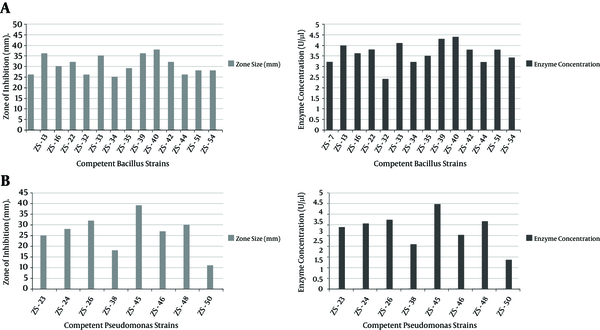
Correlative coefficience between zone of inhibition and enzyme concentration (Figure 2), depanding upon their isolation source, was also determined. Amomg Bacillus strains, ZS-39 and ZS-40 isolated from apple and mango, gave 36-mm and 38-mm zones of inhibition, and produced 4.4 ± 0.1 U/µL PG, respectively. Among Pseuodomonas strains (Figure 2B), ZS-45 isolated from orange showed maximum zone of inhibition (39 mm) and PG production (4.4 ± 0.1 U/µL) as compared to other Pseudomonas species.
Correlation Between (b/w) Zone of Inhibition and Polygalacturonase Concentration of Selected Strains: A, Bacillus; B, Pseudomonas
4.3. 16S rRNA Gene Sequencing
All competent strains selected on the basis of polygalacturinase production were characterised as Bacillus and Pseudomonas species and assigned accession numbers by NCBI. The dendrogram based on 16S rRNA gene sequence comparison showed a relationship between members of Pseudpmonas and Bacillus.
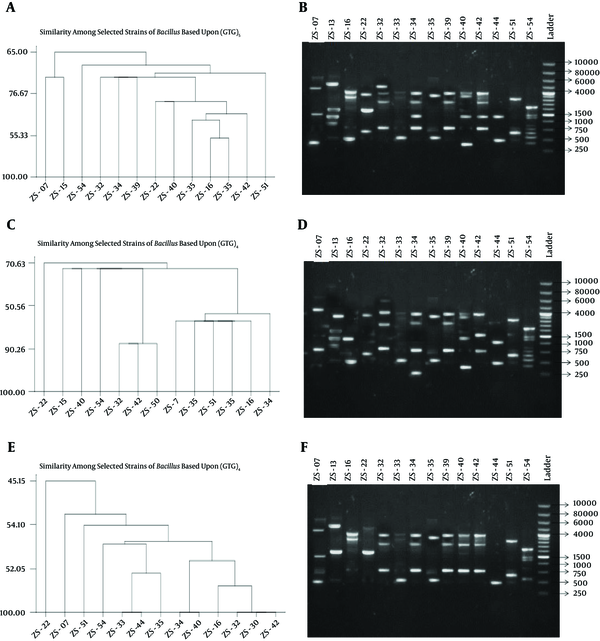
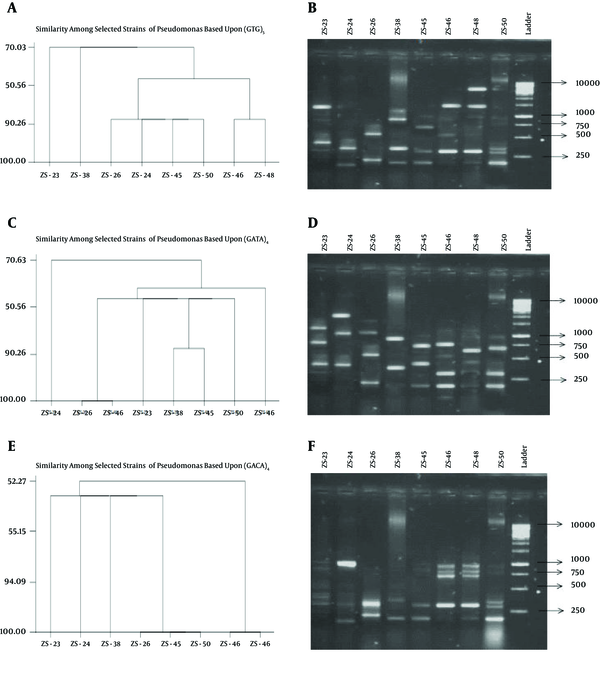
4.4. Genetic Diversity by Microsatellite DNA Fingerprinting
Genetic diversity using SSR has great advantages because of high amounts of polymorphism and presence in both coding and non-coding regions of the genome. In this study, fingerprinting of competant strains of Bacillus and Pseudomonas was done by a simple PCR technique using six sets of simple sequence repeats. Among these sets only three gave reproducible results (GACA)4, (GTG)5 and (GATA)4. Other simple repeats, which did not show any amplification, were (CA)8, (GGAT)4 and (CAA)5. polymorphism among selected strains of Bacillus and Pseudomonas was observed, and a dendrogram was constructed using Minitab 3.
Genetic Diversity in Bacillus Using Simple Sequence Repeats
Genetic Diversity in Pseudomonas Species Using Single Sequence Repeats
5. Discussion
Pectinases have been used in several industrial processes, such as fruit juice extraction, paper industry, textile processing and bioscouring of cotton fibers, retting and degumming of plant fiber, waste water treatment, coffee and tea fermentation, animal feed, purification of plant viruses, oil extraction, beverages, and wines etc. (2, 13, 14). Microorganisms including Bacillus, Pseudomonas, Erwinia, Kluyveromyces, Aspergillus, Rhizopus, Trichoderma, Penicillium, and Fusarium are competent producers of pectinases. Use of pathogens in industrial processes is not recommended so phytopathogens are being discouraged for this purpose. Mutagenic microorganisms are also discouraged due to biosafety measures. Fungi are mostly used for industrial applications, yet slow growth is the main limiting factor of fungi. For this reason, bacteria are being screened for PG production. Bacteria are easy to grow and bacterial enzymes are easy to harvest and stable, while working conditions are normal environmental conditions. Research is being conducted throughout world to screen for competent strains of bacteria for PG production. In Pakistan, little work is done on pectinase-producing microorganisms, especially bacteria. This study aimed to isolate environment friendly pectinase-producing microorganisms (Bacillus and Pseudomonas).
Among bacteria, Bacillus has been reported to produce PG (15). Polygalacturonase production by B. vallismortis (JQ990307) was similar to Bacillus spp. P 4.3 by submerged fermentation (5). In the current study, fourteen strains of Bacillus and eight strains of Pseudomonas were found competent for PG production. Molecular characterization of PG-producing strains was done by 16S rRNA sequencing. Bacillusvallismortis (ZS-40) isolated from mango produced maximum concentrations of PG, 4.4 U/µL. Orange contains good amounts of pectin (0.5% - 3.5%) and bacteria isolated from orange produced good amounts of enzyme (16). Pseudomonasaeruginosa (ZS-45), isolated from orange produced maximum enzyme activity, 4.4 U/µL. Polygalacturonase production by Enterobacter aerogenes NBO2 (18.54 U/mL) (17) was much less compared to Bacillus and Pseudomonas (4.4 U/µL each). Zeni et al. (18) reported the same results for PG production by various species of Bacillus from agro-industrial waste.
On the basis of (GACA)4, Bacillusthuringiensis (ZS-33), B. vallismortis (ZS-40) and Bacilluscereus (ZS-39) having enzyme concentration ≥ 4.0 U/µl were 70% similar to each other. In case of Pseudomonas, ZS-45 and ZS-26 (P. aeruginosa) were the most competent producers of PG were 100%, 85% and 80% similar to each other on the basis of (GACA)4, (GTG)5 and (GATA)4 respectively. Fingerprinting may be used as a molecular tool for the preliminary screening of PG producers before evaluating PG production by the plate and DNS methods. From this study, we concluded that environmental bacteria also produced excellent amounts of PG. These microorganisms are environmental friendly, don’t have fastidious nutritional requirements and are easy to grow and handle. It is easy to harvest PG than fungus as it is an extracellular product in bacterial culture. DNA motifs (GACA)4 are directly related to PG production in Bacillus and Pseudomonas.
Acknowledgements
References
-
1.
Alaña A, Gabilondo A, Hernando F, Moragues MD, Dominguez JB, Llama MJ, et al. Pectin lyase production by a Penicillium italicum strain. Appl Environ Microbiol. 1989;55(6):1612-6.
-
2.
Yadav S, Yadav PK, Yadav D, Yadav KDS. Pectin lyase: A review. Proc Biochem. 2009;44(1):1-10. https://doi.org/10.1016/j.procbio.2008.09.012.
-
3.
Tari C, Gögus N, Tokatli F. Optimization of biomass, pellet size and polygalacturonase production by Aspergillus sojae ATCC 20235 using response surface methodology. Enzyme Microb Technol. 2007;40(5):1108-16. https://doi.org/10.1016/j.enzmictec.2006.08.016.
-
4.
Silley P. The production and properties of a crude pectin lyase from Lachnospira multiparas. Lett Appl Microbiol. 1986;2(2):29-31. https://doi.org/10.1111/j.1472-765X.1986.tb01509.x.
-
5.
Soares MMCN, Silva R, Gomes E. Screening of bacterial strains for pectinolytic activity: characterization of the polygalacturonase produced by Bacillus sp. Revista de Microbiologia. 1999;30(4). https://doi.org/10.1590/s0001-37141999000400002.
-
6.
Latif Z, Sohail M. Molecular characterization of polygalacturonase producing Klebsiella and Staphylococcus species by 16S rRNA sequencing collected from rotten fruits and vegetables. Afr J Microbiol Res. 2012;6(46):7319-23.
-
7.
Hankin L, Zucker M, Sands DC. Improved solid medium for the detection and enumeration of pectolytic bacteria. Appl Microbiol. 1971;22(2):205-9. [PubMed ID: 4938100].
-
8.
Bailey MJ, Biely P, Poutanen K. Interlaboratory testing of methods for assay of xylanase activity. J Biotechnol. 1992;23(3):257-70. https://doi.org/10.1016/0168-1656(92)90074-j.
-
9.
Ausubel LM, Cramton P. Virtual power plant auctions. Utilities Policy. 2010;18(4):201-8. https://doi.org/10.1016/j.jup.2010.05.002.
-
10.
Muyzer G, de Waal EC, Uitterlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59(3):695-700. [PubMed ID: 7683183].
-
11.
Hannan A, Bajwa R, Latif Z. Status of Aspergillus niger strains for pectinases production potential. Pak J Phytopathol. 2009;21(1):77-82.
-
12.
2008.Abbreviated identification of bacteria and yeast; approved guideline-second edition. Wayne; Clinical and Laboratory Standards Institute.
-
13.
Kashyap DR, Vohra PK, Chopra S, Tewari R. Applications of pectinases in the commercial sector: a review. Bioresour Technol. 2001;77(3):215-27. [PubMed ID: 11272008].
-
14.
Jayani RS, Saxena S, Gupta R. Microbial pectinolytic enzymes: A review. Proc Biochem. 2005;40(9):2931-44. https://doi.org/10.1016/j.procbio.2005.03.026.
-
15.
Swain MR, Kar S, Ray RC. Exo-polygalacturonase production by Bacillus subtilis CM5 in solid state fermentation using cassava bagasse. Braz J Microbiol. 2009;40(3):636-48. [PubMed ID: 24031409]. https://doi.org/10.1590/S1517-838220090003000028.
-
16.
Voragen AGJ, Schols HA, Pilnik W. Determination of the degree of methylation and acetylation of pectins by h.p.l.c. Food Hydrocolloids. 1986;1(1):65-70. https://doi.org/10.1016/s0268-005x(86)80008-x.
-
17.
Darah I, Nisha M, Lim SH. Polygalacturonase production by calcium alginate immobilized Enterobacter aerogenes NBO2 cells. Appl Biochem Biotechnol. 2015;175(5):2629-36. [PubMed ID: 25547814]. https://doi.org/10.1007/s12010-014-1447-4.
-
18.
Zeni J, Cence K, Grando CE, Tiggermann L, Colet R, Lerin LA, et al. Screening of pectinase-producing microorganisms with polygalacturonase activity. Appl Biochem Biotechnol. 2011;163(3):383-92. [PubMed ID: 20669053]. https://doi.org/10.1007/s12010-010-9046-5.