Abstract
Background:
Methicillin-resistant Staphylococcus aureus (MRSA) is a bacterial pathogen frequently isolated in both hospital and community environments. Methicillin-resistant Staphylococcus aureus is considered a major nosocomial pathogen that causes severe morbidity and mortality.Objectives:
The main objective of this study was to determine the genotypes of MRSA strains isolated from the nares of hospitalized and community patients in Kermanshah Hospital, western Iran, by PCR-restriction fragment length polymorphism (PCR-RFLP).Materials and Methods:
Of 1387 patients, 1217 patients were screened for more than 48 hours after admission in hospital wards and 170 patients were screened in the hemodialysis unit of Kermanshah Hospital, which is the largest hospital in western Iran. S. aureus was identified by standard biochemical tests, including colonial morphology, production of coagulase, and DNase and the API20 Staph test. Methicillin-resistant Staphylococcus aureus was identified by the Oxacillin strip test.Results:
In total, 258 S. aureus isolates were recovered from 1387 samples, of which 96 isolates were MRSA, 82 were hospital acquired, and 14 were community acquired. Digestion of the aro A gene revealed only one distinctive RFLP pattern in the 258 isolates.Conclusions:
Methicillin-resistant Staphylococcus aureus is an increasingly common cause of nosocomial infections. Our results are in agreement with those of other studies reporting that a few specialized clones are responsible for most cases of MRSA nasal carriage. In this study, MRSA strains isolated from different wards of hospital were closely related when analyzed by coagulase gene typing. Identifying patients colonized with MRSA during hospitalization and rapidly typing them with these methods may facilitate detection of outbreaks and prevention of the spread of organisms in hospitals.Keywords
Restriction Fragment Length Polymorphisms Methicillin-Resistant Staphylococcus aureus Coa gene aroA gene Staphylococcus aureus
1. Background
Staphylococcus aureus could be considered a major cause of human diseases, particularly in the hospital setting. It is one of the most feared pathogens because of its ability to cause overwhelming sepsis and death (1). Methicillin-resistant Staphylococcus aureus (MRSA) is a bacterial pathogen frequently isolated and presented in community and hospital environments (2). Since the first report of MRSA in 1961, it has been considered an important pathogen in hospitalized patients that causes severe morbidity and mortality worldwide and is a major problem for public health and clinical settings (3, 4). Although hospital-acquired (HA) MRSA infections were initially reported in the 1980s, immediately after the introduction of methicillin in the hospital settings and they have continued to increase in last year’s (5).
The potential for HA infections increases when patients with unidentified community colonization are placed in healthcare-associated environments. In an active surveillance program, a patient is screened for carrying a selected organism, regardless of whether or not the patient is exhibiting signs and symptoms of infection at that time (6). The rate of nasal carriage of S. aureus isolates was found to vary from 10% to up 90% (7-9). Molecular typing plays an important role in epidemiological studies. Methods such as polymerase chain reaction–restriction fragment length polymorphism (PCR-RFLP) typing are alternatives to more expensive techniques, such as pulsed-field gel electrophoresis (PFGE) (9). Pulsed-field gel electrophoresis is a labor-intensive and time-consuming technique and can be performed only in reference research laboratories with technically demanding but expensive than the present typing techniques such as coagulase gene restriction profile (CRP) analysis and require only optimized and standard protocols (10).
A PCR-based typing systems, such as rep-PCR, PCR-RFLP analysis of the coa and aroA gene would be appropriate for detection and typing of S. aureus isolates, because these are simple, inexpensive, rapid, reproducibility and easy in applicable settings and also ideal typing method for analysis of large numbers of Methicillin-resistant Staphylococcus aureus isolates in many outbreaks result from these organisms (11). The aroA gene encodes the enzyme 5-enolpyruvyl-shikimate 3-phosphate synthase. S. aureus isolates can be confirmed with PCR amplification of the aroA gene (12). On the other hand, coagulase is produced by all isolates of S. aureus. Coagulase (coa) gene typing has been used to identify and compare S. aureus genotypes, as it is a simple, specific, discriminatory, and reproducible method for the typing of S. aureus isolated from plain sources (13, 14).
2. Objectives
The main objective of this study was to determine genotypes of MRSA strains isolated from the nares of hospitalized and community patients in Kermanshah Hospital, western Iran, by PCR-RFLP.
3. Materials and Methods
This cross-sectional study was conducted from October 2012 to August 2013, in 1387 patients at the Imam Reza hospital, which is a major referral hospital in western Iran with 514 tertiary care beds and 10 health wards, including Emergency, Pediatrics, Internal Medicine, Intensive Care Unit (ICU) Infectious diseases, Critical Care Unit, and Pediatrics. In total, 1217 patients in different wards were screened more than 48 hour after admission for HA isolates, that is, for nosocomial infections defined as infections noted 48 hour after admission. Another 170 patients in the hemodialysis unit were screened for community-acquired isolates without gathering any admission history. Exclusion criteria for patient enrollment included the following items: antibiotic therapy in the previous week, hospitalization during the previous month, history of Staphylococcus infections, and known immune deficiency.
Nasal samples (n = 1387) were taken using cotton sterile swabs moiled with sterile saline. Sampling was performed using methods described by Mohajeri et al. (8). All swabs were transported in the Amies medium and collected by the same researcher during the study. The swabs were directly inoculated on Mannitol salt agar (Merck, Germany). Each distinctive morphotype of the mannitol-fermenting colony was selected from the plate, subcultured on a blood agar plate (Merck, Germany), and incubated at 37ºC in an incubator under humidified conditions. S. aureus isolates were identified by standard phenotypic tests such as colonial morphology, coagulase production, and DNase and the API 20 staph test (bioMerieux, France). MRSA was identified by the Oxacillin strip test (Mast, UK) (15).
3.1. DNA Extraction
Staphylococcus aureus genomic DNA was extracted using the standard phenol–chloroform method. PCR for the aroA and coa genes and RFLP for aroA were performed according to the method described by Dastmalchi Saei et al. (Table 1), with minor modifications (16). After optimizing PCR reactions for the amplification of the aroA gene, the reaction was carried out in a DNA thermal cycler (BioRad C1000, USA) for obtaining a single amplicon with an anticipated size of 1153 bp. Each PCR reaction was carried out in a final volume of 50 µL reaction containing 2 µL of template DNA, 0.5 mM of each primer, the four deoxynucleoside triphosphates (each at 200 mM), PCR buffer 10 × 1.5 mM of MgCl2, and 1.5 Units of TaqDNA polymerase (Sinaclone, Iran).
Staphylococcus aureus ATCC 25923 was used as the positive control for the aroA and coa genes and sterile water was used as the negative control. Amplified products were detected by electrophoresis at 60 V for 20 minutes and at 45 V for 70 minutes and photographed by Gel Doc (Bio-Rad XR+, Hercules, CA, USA). When the aroA gene was amplified, 10 µL of the PCR product of this reaction was incubated with 16 µL of sterile water, 3 µL of 10 × buffer, and 1 µL (20 U/μl) of HaeIII restriction enzyme (Fermentas, EU) at 37ºC for 2.5 hours. The digested PCR product was electrophoresed on a 1.5% agarose gel at 80 V for 60 minutes. Coagulase gene typing was performed as described by Dastmalchi Saei et al. with some modifications (16). The method is based on the heterogenicity of a region including tandem repeats with 81-bp size at the 3’ end of the coa gene. After confirming the amplification of the variable region of the coa gene, 10 µL of the PCR product of this gene was incubated with 16 µL of sterile water, 3 µL of 10 × buffer, and 1 µL (20 U/μl) of HaeIII restriction enzyme (Fermentas, EU) at 37ºC for 2.5 hour. The digested PCR products were electrophoresed on a 1.5% agarose gel at 80 V for 60 minutes. The products were stained with ethidium bromide and the images were obtained by a gel documentation (BioRad XR +, Hercules, CA, USA) system.
Primers to aroA and Coa Used in Polymerase Chain Reaction
| Primer Name | Sequence | Product Size (base pair) |
|---|---|---|
| aroA F1 | 5ʹ-AAG GGC GAA ATA GAA GTG CCG GGC-3ʹ | 1153 |
| aroA R2 | 5ʹ-CAC AAG CAA CTG CAA GCA T-3ʹ | |
| coa1 | 5ʹ-ATA GAG ATG CTG GTA CAG G-3ʹ | 850 |
| coa2 | 5ʹ-GCT TCC GAT TGT TCG ATG C-3ʹ |
4. Results
In this study, 258 S. aureus isolates were recovered from 1387 samples: 96 isolates were MRSA, which included 82 HA isolates and 14 community-acquired isolates. The 96 isolates were collected from different wards: 6 (6.2%), ICU; 11 (11.5%), surgery; 20 (20.8%), internal medicine; 7 (7.3%), gynecology; 5 (5.2%), infection; 1 (1%), heart; 21 (21.87%), pediatrics; and 25 (26%), hemodialysis. Eight age groups were defined (Table 2). The aroA gene was successfully amplified in all MRSA isolates tested (Figure 1). Digestion of the aroA gene yielded one distinctive RFLP pattern. All the isolates generated approximately 300- and 850-bp bands (Figure 1). In contrast, PCR amplification of coa produced bands of five different sizes, ranging from 490 bp to 810 bp (Figure 2). RFLP analysis of the coa gene showed five distinctive patterns (Figure 3). Table 3 shows the genotype distribution of HA and community-acquired isolates, and Table 4 lists the patterns seen within the different wards in our hospital. Table 3 also shows coa gene typing and PCR-RFLP patterns detected using restriction enzyme AluI.
Distribution of Patients by Age
| Age Group | Frequency, % |
|---|---|
| < 10 | 20 (20.8) |
| 10 - 19 | 3 (3.1) |
| 20 - 29 | 5 (5.2) |
| 30 - 39 | 9 (9.4) |
| 40 - 49 | 7 (7.3) |
| 50 - 59 | 20 (20.8) |
| 60 - 69 | 16 (16.7) |
| ≥ 70 | 16 (16.7) |
| Total | 96 (100.0) |
Agarose gel Electrophoresis of the aroA Gene PCR Amplification Products
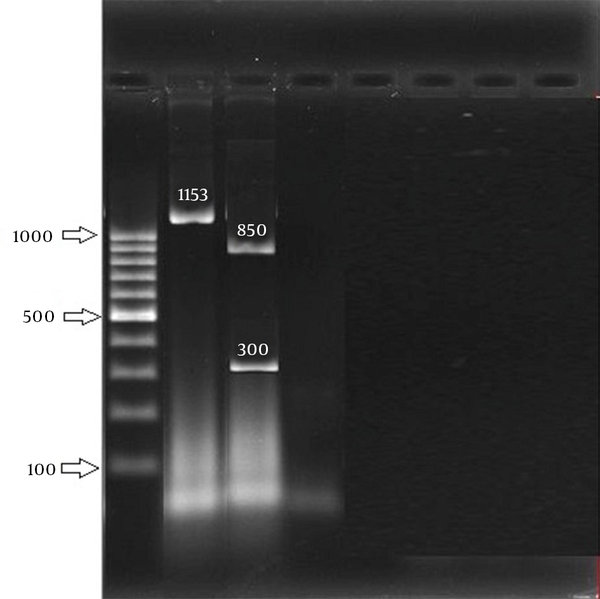
Agarose gel Electrophoresis of the coa Gene PCR Amplification Products and the PCR Amplified aroA Gene
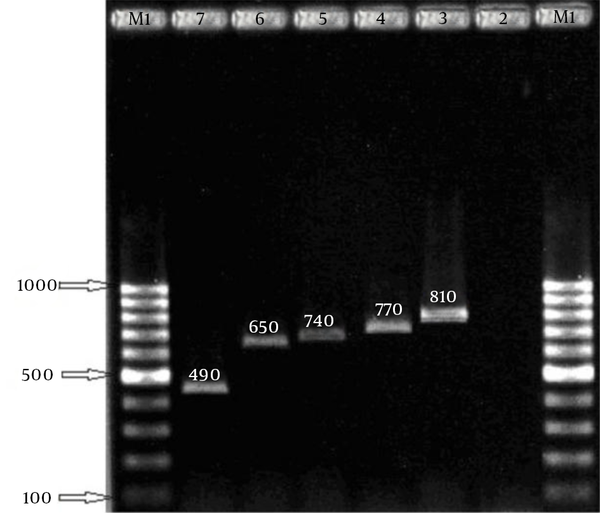
PCR-amplified coa Gene Digested (RFLP) With HaeIII Restriction Endonuclease
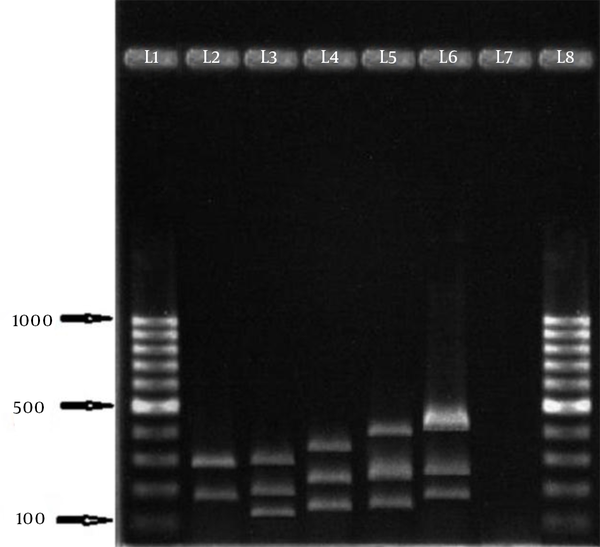
Genotypes Distribution of Hospital-acquired and Community-acquired Isolates and Their RFLP Patterns
| Genotype Code | PCR Product, bp | No. of Isolates | No. of Hospital-acquired Isolates | No. of Community-acquired Isolates | RFLP Patterns, bp |
|---|---|---|---|---|---|
| I | 810 | 15 | 6 | 9 | 360, 260, 190 |
| II | 770 | 13 | 9 | 4 | 350, 260, 160 |
| III | 740 | 29 | 25 | 4 | 320, 260, 160 |
| IV | 650 | 27 | 22 | 5 | 300, 200, 190 |
| V | 490 | 12 | 9 | 3 | 300, 190 |
Distribution of HaeIII Restriction Patterns Within Different Hospital Wards
| Ward | No. of Isolates | Genotype Code | ||||
|---|---|---|---|---|---|---|
| I | II | III | IV | V | ||
| ICU | 6 | 1 | 1 | 1 | 3 | 0 |
| Surgery | 11 | 1 | 2 | 4 | 2 | 2 |
| Internal medicine | 20 | 0 | 4 | 5 | 6 | 5 |
| Gynecology | 7 | 1 | 1 | 2 | 3 | 0 |
| Infection | 5 | 0 | 1 | 0 | 4 | 0 |
| Heart | 1 | 0 | 0 | 1 | 0 | 0 |
| Pediatrics | 5 | 1 | 0 | 3 | 1 | 0 |
| Infants | 16 | 2 | 0 | 9 | 3 | 2 |
| Hemodialysis | 25 | 9 | 4 | 4 | 5 | 3 |
5. Discussion
As the model pathogen, we selected MRSA because this multidrug-resistant nosocomial pathogen has previously been studied genetically worldwide (17, 18). Methicillin-resistant Staphylococcus aureus isolates are suitable as the best model for investigating infection control and outbreak in hospitals and community (12). Methicillin-resistant Staphylococcus aureus is an increasingly common cause of nosocomial outbreaks with several morbidity and mortality worldwide, and more than 50% of all S. aureus diseases occurred in some part of hospitals. Another study reported that detection of hospital and community isolates of MRSA in nasal samples and their treatment may be an important modality in the prevention of infections by these isolates (19, 20).
In this regard, molecular typing provides valuable information on the genetic background of MRSA isolates. These techniques have been useful for differentiation of MRSA isolates and are further used as tools for epidemiological purposes (10). In the present study, PCR products of five different sizes were obtained from amplification of the coa gene. Similarly, studies by other researchers in different parts of world (12) conducted using the same primer pairs also showed distinct coagulase gene types in some isolates. This variation and polymorphism among some isolates of S. aureus is unclear, but some studies attribute it to the insertion or deletion mutations that occur at the 3ʹ end of the coa gene; these mutations change the coa gene size and also change the antigenic properties of the coagulase enzyme in serological tests. This region of the coa gene may play an important role in the antigenic variation of coagulase and its escape from the inhibitory effect of anticoagulase agents (17).
Our results and those of other studies have shown that some specialized clones are responsible for the outbreak, most cases of MRSA nasal carriage, and skin colonization (16, 21). The 3′ end of the coa gene contains a series of tandem repeats 81-bp in size, which is distinctive among S. aureus isolates within their number or in the location of AluI restriction sites. The production of coa is important for the future of epidemiology typing and molecular identification of S. aureus (13). Classification of S. aureus isolates based on the coa gene is a simple and accurate method for molecular typing (13, 18, 22). Detection and isolation of carriers is crucial for controlling the spread of nosocomial infection with MRSA isolates in a hospital setting. During nosocomial outbreaks, a rapid MRSA screening test based on molecular epidemiology typing can be used for detecting the DNA of epidemic strains in colonized sites of patients. Potential targets for genotyping of MRSA isolates based on the detection of conserved sequences include specific nucleotide sequences such as 81-bp tandem repeats in the 3′ end of the coa gene, which is considered effective in identifying outbreaks. In this study, MRSA strains isolated in the different wards of the hospital were closely related when analyzed by aroA gene typing.
In the present study, HaeIII enzyme, which has been used in previous studies (23, 24) for generating RFLP patterns of the coa gene of S. aureus, did not digest all PCR products. RFLP analysis for the coa gene showed five distinctive patterns in these isolates. A possible reason for this finding might be alterations in the polymorphic repeated region of the coa gene because of point mutations (23, 24). As mentioned before, isolates with 740- and 650-bp PCR products were the predominant types, and interestingly all isolates with the 740- and 650-bp PCR products yielded the same RFLP pattern, indicating no heterogeneity in HaeIII recognition sites among this predominant type (25). Aarestrup et al. reported that coa gene polymorphism is useful for classifying S. aureus strains (26), because of its high typability, reproducibility, and good discriminatory power, and also because it can be used in epidemiological investigations and for molecular typing of S. aureus isolated from different origins (27). In contrast to our findings in human isolates, Dastmalchi Saei et al., in their study in northwest Iran, observed that 58 S. aureus isolates from bovine mastitis were distributed over nine RFLP different patterns with 490- to 850-bp fragments from digestion with HaeIII enzyme (16). Each of these patterns differed in frequency, with the highest frequency observed for pattern I (37%) followed by pattern VIII (24%).
5.1. Conclusion
Colonization by a microorganism, an essential step leading to infection with that microorganism, may persist for months or years. Patients with MRSA colonization serve as a reservoir for MRSA transmission in hospitals. Nasal carriage of S. aureus is a substantial source of human infections in some wards of hospitals. Identifying patients colonized with MRSA during hospitalization and rapidly typing them with the methods presented in this paper may facilitate detection of outbreaks and prevention of the spread of the organism in hospitals. Our results showed that RFLP types may differ between hospitalized and community patients in hospitals.
Acknowledgements
References
-
1.
Mahon CR, Lehman DC, Manuselis G. Diagnostic microbiology. 3th ed. Philadelphia: Saunders; 2007.
-
2.
Moon JS, Lee AR, Kang HM, Lee ES, Kim MN, Paik YH, et al. Phenotypic and genetic antibiogram of methicillin-resistant staphylococci isolated from bovine mastitis in Korea. J Dairy Sci. 2007;90(3):1176-85. [PubMed ID: 17297092]. https://doi.org/10.3168/jds.S0022-0302(07)71604-1.
-
3.
Lamaro-Cardoso J, Castanheira M, de Oliveira RM, e Silva SA, Pignatari AC, Mendes RE, et al. Carriage of methicillin-resistant Staphylococcus aureus in children in Brazil. Diagn Microbiol Infect Dis. 2007;57(4):467-70. [PubMed ID: 17240106]. https://doi.org/10.1016/j.diagmicrobio.2006.10.008.
-
4.
Yinduo J. Methods in Molecular Biology. In: Yinduo J, editor. Methicillin-resistant Staphylococcus aureus (MRSA) protocols. New York: Humana Press; 2007. 391 p. https://doi.org/10.1007/978-1-62703-664-1.
-
5.
Mohanasoundaram KM, Lalitha MK. Comparison of phenotypic versus genotypic methods in the detection of methicillin resistance in Staphylococcus aureus. Indian J Med Res. 2008;127(1):78-84.
-
6.
Cesur S, Cokca F. Nasal carriage of methicillin-resistant Staphylococcus aureus among hospital staff and outpatients. Infect Control Hosp Epidemiol. 2004;25(2):169-71. [PubMed ID: 14994946]. https://doi.org/10.1086/502371.
-
7.
Eveillard M, Martin Y, Hidri N, Boussougant Y, Joly-Guillou ML. Carriage of methicillin-resistant Staphylococcus aureus among hospital employees: prevalence, duration, and transmission to households. Infect Control Hosp Epidemiol. 2004;25(2):114-20. [PubMed ID: 14994935]. https://doi.org/10.1086/502360.
-
8.
Mohajeri P, Izadi B, Rezaei M, Farahani A. Frequency Distribution of Hospital-Acquired MRSA Nasal Carriage Among Hospitalized Patients in West of Iran. Jundishapur J Microbiol. 2013;6(6). https://doi.org/10.5812/jjm.9076.
-
9.
Godwin PG, Choyce A, McCarthy S. The prevalence of MRSA carriage measured over five years in a District General Hospital. J Hosp Infect. 2001;47(1):73-5. [PubMed ID: 11161906]. https://doi.org/10.1053/jhin.2000.0866.
-
10.
Chiou CS, Wei HL, Yang LC. Comparison of pulsed-field gel electrophoresis and coagulase gene restriction profile analysis techniques in the molecular typing of Staphylococcus aureus. J Clin Microbiol. 2000;38(6):2186-90. [PubMed ID: 10834974].
-
11.
Shittu AO, Lin J. Antimicrobial susceptibility patterns and characterization of clinical isolates of Staphylococcus aureus in KwaZulu-Natal province, South Africa. BMC Infect Dis. 2006;6(125):1-13.
-
12.
Hookey JV, Richardson JF, Cookson BD. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J Clin Microbiol. 1998;36(4):1083-9. [PubMed ID: 9542942].
-
13.
Goh SH, Byrne SK, Zhang JL, Chow AW. Molecular typing of Staphylococcus aureus on the basis of coagulase gene polymorphisms. J Clin Microbiol. 1992;30(7):1642-5. [PubMed ID: 1352784].
-
14.
Vimercati C, Cremonesi P, Castiglioni B, Pisoni G, Boettcher PJ, Stella A, et al. Molecular typing of Staphylococcus aureus isolated from cows, goats and sheep with intramammary infections on the basis of gene polymorphisms and toxins genes. J Vet Med B Infect Dis Vet Public Health. 2006;53(9):423-8. [PubMed ID: 17062119]. https://doi.org/10.1111/j.1439-0450.2006.00980.x.
-
15.
Farahani A, Mohajeri P, Gholamine B, Rezaei M, Abbasi H. Comparison of different phenotypic and genotypic methods for the detection of methicillin-resistant Staphylococcus aureus. N Am J Med Sci. 2013;5(11):637-40. [PubMed ID: 24404541]. https://doi.org/10.4103/1947-2714.122305.
-
16.
Dastmalchi Saei H, Ahmadi M, Mardani K, Batavani RA. Molecular typing of Staphylococcus aureus isolated from bovine mastitis based on polymorphism of the coagulase gene in the north west of Iran. Vet Microbiol. 2009;137(1-2):202-6. [PubMed ID: 19195799]. https://doi.org/10.1016/j.vetmic.2009.01.001.
-
17.
Momtaz H, Tajbakhsh H, Rahimi R, Momeni M. Paediatrics & Child Health 2005. 23th ed. Singapore: Nations Communities in Canada; 2004. p. 223-30.
-
18.
Silva ER, Boechat JU, Silva N. Coagulase gene polymorphism of Staphylococcus aureus isolated from goat mastitis in Brazilian dairy herds. Lett Appl Microbiol. 2006;42(1):30-4. [PubMed ID: 16411916]. https://doi.org/10.1111/j.1472-765X.2005.01799.x.
-
19.
Erami M, Soltani B, Taghavi Ardakani A, Moravveji A, Haji Rezaei M, Soltani S, et al. Nasal Carriage and Resistance Pattern of Multidrug Resistant Staphylococcus aureus Among Healthy Children in Kashan, Iran. Iran Red Crescent Med J. 2014;16(9). eee21346. [PubMed ID: 25593734]. https://doi.org/10.5812/ircmj.21346.
-
20.
Ghadiri K. Nasal carriage rate of community-and hospital-acquired methicillin-resistant staphylococcus aureus in children, Kermanshah, Iran. Archives Clin Infect Dis. 2012;6(3):117-20.
-
21.
Reza TS, Malahat A, Dastmalchi SH. Restriction fragment length polymorphism genotyping of human Staphylococcus aureus isolates from two hospitals in urmia region of iran using the coa gene. Jundishapur J Microbiol. 2012;2012(2):416-20. https://doi.org/10.5812/jjm.3522.
-
22.
Rodrigues da Silva E, da Silva N. Coagulase gene typing of Staphylococcus aureus isolated from cows with mastitis in southeastern Brazil. Can J Vet Res. 2005;69(4):260-4. [PubMed ID: 16479723].
-
23.
Ishino K, Tsuchizaki N, Ishikawa J, Hotta K. Usefulness of PCR-restriction fragment length polymorphism typing of the coagulase gene to discriminate arbekacin-resistant methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 2007;45(2):607-9. [PubMed ID: 17166956]. https://doi.org/10.1128/JCM.02099-06.
-
24.
Karahan M, Cetinkaya B. Coagulase gene polymorphisms detected by PCR in Staphylococcus aureus isolated from subclinical bovine mastitis in Turkey. Vet J. 2007;174(2):428-31. [PubMed ID: 16901735]. https://doi.org/10.1016/j.tvjl.2006.05.016.
-
25.
Jariyasethpong T, Tribuddharat C, Dejsirilert S, Kerdsin A, Tishyadhigama P, Rahule S, et al. MRSA carriage in a tertiary governmental hospital in Thailand: emphasis on prevalence and molecular epidemiology. Eur J Clin Microbiol Infect Dis. 2010;29(8):977-85. [PubMed ID: 20509037]. https://doi.org/10.1007/s10096-010-0954-7.
-
26.
Aarestrup FM, Dangler CA, Sordillo LM. Prevalence of coagulase gene polymorphism in Staphylococcus aureus isolates causing bovine mastitis. Can J Vet Res. 1995;59(2):124-8. [PubMed ID: 7648524].
-
27.
Raimundo O, Deighton M, Capstick J, Gerraty N. Molecular typing of Staphylococcus aureus of bovine origin by polymorphisms of the coagulase gene. Vet microbiol. 1999;66(4):275-84. https://doi.org/10.1016/S0378-1135(99)00020-6.