Abstract
Background:
Pseudomonas aeruginosa is a Gram negative ubiquitous opportunistic organism and one of the more problematic drug-resistant pathogens encountered today.Objectives:
The aims of the current multicenter research were to assess antibiotic resistance profiles, carbapenemase production, and detection of antibiotic resistance IMP gene as well as virulence factors genes including exoA, algD, lasB, and plcH among the clinical isolates of P. aeruginosa.Methods:
A total of 80 nonduplicate isolates of P. aeruginosa were recovered from inpatients. Bacterial identification was done by standard diagnostic tests. Species was confirmed by detection of the exoA gene using the PCR technique. Antimicrobial susceptibility test was performed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. Carbapenemase production among the isolates was determined by the modified Hodge test. Virulence genes were detected by PCR.Results:
A total of 42 (52.5%) isolates recovered from wound specimens. Colistin was the most effective antibiotic against isolates (97.5% isolates were susceptible) and levofloxacin was the least effective drug (67.5% isolates were resistant). The most common antibiotic resistance pattern was CIPR-CPMR-GEMR (47 isolates). In total, 47 (58.75%) isolates were identified as multidrug resistance (MDR), while 30% of isolates were carbapenemase producer (MHT+). Among studied isolates, plcH+ and lasB+ genotypes (100% isolates) were the most common virulence gene patterns. Of 80 P. aeruginosa isolates, 39 (48.75%) showed algD+, plcH+, lasB+, and IMP+ genotype. The blaIMP resistance gene was detected in all MHT positive and MDR isolates.Conclusions:
In our study, the emergence of potentially highly pathogenic and carbapenem-resistant strains in joining with a MDR phenotype is alarming, as a feasible outcome would be a severe clinical result concomitant with critical restrictions in antibiotic therapy.Keywords
Hospital Pseudomonas aeruginosa Multidrug Resistance Virulence Factors
1. Background
Pseudomonas aeruginosa is a Gram negative, bacillary shape, non-spore forming, ubiquitous opportunistic bacterium present in many various environmental settings in different geographical regions, and it can be isolated from diverse living sources, such as humans, animals, and plants (1, 2). The potency of this organism to tolerate a variety of unsuitable physical conditions and to survive on minimal nutritional requirements has allowed this bacterium to persist in both hospital wards and community (2, 3). Pseudomonas aeruginosa is a member of the normal microbial flora in humans. Representative colonization rates for specific sites in human body are 0% to 3.3% for the nasal mucosa, 0% to 2% for skin, 2.6% to 24% for stool, and 0% to 6.6% for the throat specimens (1, 4).
Colonization rates may exceed 50% during hospitalization, especially among persons who have experienced trauma to or a breach in mucosal barriers or cutaneous by severe burns, mechanical ventilation, catheters, surgery, or tracheostomy (5). Disruption in the normal microbial flora as a result of prolonged antibiotic therapy has also been shown to increase colonization by P. aeruginosa; also immunocompromised individuals with deficiency in immune system have higher risks for colonization by this bacterium (6). Pseudomonas aeruginosa causes several different infections including endocarditis, pneumonia, malignant otitis externa, bacteremia, and also, gastrointestinal tract, skin and soft tissue, skeletal, eye, and burn infections (7, 8). Pseudomonas aeruginosa was the first leading cause of burn infections and the 3rd most common cause of infection in the urinary tract (7% to 11%) (1, 7).
The rapid increase of multidrug-resistant P. aeruginosa (MDRPA) isolates in clinical settings worldwide has resulted in an increased mortality rate (9, 10). Several mechanisms are involved in P. aeruginosa resistance to antimicrobial agents, such as carbapenemase production. In addition, the propagation of various classes of carbapenem hydrolyzing bacterial enzymes has been rising worldwide (11, 12). All types of transferable carbapenemases have been identified in P. aeruginosa strains in different geographical regions. Among these enzymes, the metallo-beta-lactamases (MBLs) are considered as the most clinically important for P. aeruginosa. Most MBLs encoding genes are located on mobile genetic elements (10, 13). Furthermore, resistance genes for other antibiotic classes can be present in the same mobile genetic elements contributing in the growth and propagation of MDR isolates. IMP-type MBLs is spread worldwide and in different geographical areas after their first detection in Japan (14). Pseudomonas aeruginosa have different virulence factors.
Many extracellular pathogenic determinants have been shown to be regulated by a complex regulatory circuit including quorum sensing pathway (15). Some of the most important virulence factors of P. aeruginosa are Exotoxin A, alginate, and haemolytic phospholipase C. Exotoxin A (ExoA) is a 66 Kd protein, which acts as a major virulence factor of P. aeruginosa (1, 16-19). The production of mucoid colonies by P. aeruginosa strains composed of alginates, involving algD genes, protects the organism from antimicrobials and the host’s immune system response, and therefore, contributes to chronic inflammation of pulmonary system. Moreover, the phospholipids included in pulmonary system surfactants may be degraded by 2 phospholipases C encoded by the plcH gene (20). These virulence markers are responsible for colonization and adhesion of bacterium to cell and destroy of host tissue.
2. Objectives
The aims of the present multicenter study were to assess antibiotic resistance profiles, carbapenemase production and detection of antibiotic resistance IMP gene as well as virulence factors genes including exoA (exotoxin A encoding gene), algD (alginate encoding gene), and plcH (haemolytic phospholipase C processor encoding gene), among the clinical isolates of P. aeruginosa recovered from patients hospitalized in main referral hospitals in the cities of Shiraz and Yasuj, southwest of Iran.
3. Methods
3.1. Ethics Statement
This work was performed according to University ethics committee code number IR.YUMS.REC.1395.74.
3.2. Specimen Collection
In this multi-center study, from April 2015 to March 2016, a total of 80 nonduplicate isolates of P. aeruginosa were recovered from inpatients hospitalized in 2 main referral hospitals in the cities of Shiraz and Yasuj. Samples were taken by expert technicians from hospitalized patients admitted to burn, general internal medicine, ICU, pediatric/neonatal, surgery, and the gynecology ward. The sampling method from the wounds included swabs collected from clinically deep areas of the burn wounds when clinical manifestations of wound infection emerged. Urine culture was prepared from patients with suspected bacteriuria and inoculated into MacConkey medium. Wound specimens were sent to clinical microbiology laboratory during 1 hour of collections.
3.3. Bacterial Identification
The collected specimens were cultured on blood agar and MacConkey agar (Merck, Germany), and incubated for 24 hours at 35°C ± 2. Primary bacterial identification was done by standard diagnostic tests. Briefly, the overnight pure growth of the organisms on MacConkey agar plates was checked on the basis of Gram staining, colonial morphology, and lactose fermentation. The isolated colonies were final identified by oxidase, catalase, motility, triple sugar iron agar (TSI) inoculation, citrate utilization, gelatin liquefaction, urease production, nitrate reduction, alkaline protease production, oxidative-fermentative reaction, indole, H2S production, and hemolysin tests. Finally, a species was confirmed by detection of exoA gene using the PCR technique, as described in the following. Prototype strain P. aeruginosa ATCC 27853 was applied as quality control strain throughout this study. The pure bacterial colonies were inoculated onto the medium containing 1.5 mL of sterile Tryptic Soy broth (TSB) (Merck, Germany) mixed with glycerol (20%) and stored at -20°C for further investigation.
3.4. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility test (AST) was performed using the disk diffusion (DD) break point assay, according to clinical and laboratory standards institute guidelines (CLSI, 2010) (21). All antibiotic disks were purchased from Mast Co. Ltd, UK and used as per manufacturer’s instructions. The following antibiotic disks were used for antimicrobial susceptibility testing: cefepime (CPM, 30 µg), ceftazidime (CAZ, 30 µg), ticarcilin (TC, 75 µg), carbenicilin (PY, 100 µg), piperacilin (PRL, 100 µg), piperacillin/tazobactam (PTZ, 110 µg), aztreonam (ATM, 30 µg), imipenem (IMI, 10 µg), meropenem (MEM, 10 µg), doripenem (DOR, 10 µg), amikacin (AK, 30 µg), tobramycin (TN, 10 µg), gentamicin (GM, 10 µg), ciprofloxacin (CIP, 5 µg), levofloxacin (LEV, 5 µg), and polymyxin B (PB, 300 U). P. aeruginosa ATCC 27853 was used as a standard strain.
3.5. Detection of Multidrug-Resistant Isolates
Multidrug-resistant P. aeruginosa strains were defined as resistant to at least 3 of the following drugs belonging to several class of antibiotics: ceftazidime, imipenem, gentamicin, and ciprofloxacin by using the disk diffusion method. The data were interpreted according to the CLSI (2010) protocols (21).
3.6. Identification of Carbapenemase Producer Strains by Modified Hodge Test (MHT) Phenotypic Method
Carbapenemase production among the isolates was determined by modified Hodge test (MHT), according to the CLSI guidelines (21). The MHT was performed for carbapenem resistant isolates. A 0.5 McFarland dilution of the Escherichia coli ATCC 25922 in 5 mL of normal saline was prepared. A 1:10 dilution was streaked as lawn on to a Mueller Hinton agar (MHA) (Merck Co., Germany) plate. A 10 µg ertapenem antibiotic susceptibility disk was placed in the center of the MHA plate. Test isolate was streaked in a straight line from the edge of the disk to the edge of the plate. The MHA plate was incubated in ambient air for 16 to 24 hours at 35 ± 2°C. After 24 hours of incubation, MHT positive test showed a clover leaf-like indentation of the Escherichia coli 25922 growing along the test organism growth streak within the disk diffusion zone. MHT negative test displayed no growth of the E. coli 25922 along the test isolate growth streak within the disk diffusion. MHT positive Klebsiella pneumoniae ATCC1705 and MHT negative P. aeruginosa ATCC 27853 were applied as quality control (QC) with each batch of the assay.
3.7. Confirmation of Pseudomonas aeruginosa Species by Molecular Method
Polymerase chain reaction (PCR) was performed to further confirm isolates at the species level by using exoA gene encoding exotoxin A specific to P. aeruginosa. All bacterial isolates, identified as presumptive P. aeruginosa, by morphological and biochemical tests, were further confirmed by species-specific PCR to detect the exoA gene. The bacterial cells were cultured overnight on Mueller-Hinton agar. The whole genomic DNA was extracted from single colonies using the boiling method and used as a template for PCR amplification. The primer sequences and the expected sizes of amplicons are presented in Table 1. PCR mixture consisted of 2.5 µL of PCR buffer, 2.5 units of Taq DNA polymerase (Thermo Scientific), 10 pmol of each primer, 1 µL of dNTP mix (Thermoscientific Co.,), 2 µL of DNA template, and molecular grade distilled water was used to adjust the reaction volume to 25 µL.
DNA amplification was conducted in temperature gradient thermal cycler (Eppendorf 96-well, Germany). The PCR program procedure was as follows; pre-denaturation at 95°C for 5 minutes (one cycle), 34 cycles (denaturation at 95°C for 30 seconds, annealing at 58°C for 30 seconds, and extension at 72°C for 30 seconds), followed by 1 cycle of final extension at 72°C for 5 minutes. 5 µL of PCR products was subject to gel electrophoresis using 1.5% agarose gel (Merck, Germany), prepared in TBE (Tris-Borate-EDTA) buffer for 1 hour, stained with UV illuminating dye (Gel Red) and visualized by UV-gel documentation system (Kodak Gel Logic 200, USA). DNA ladder (size range 100 - 1500 bp) was used to detect the size of the expected bands.
Nucleotide Sequences of Primers and Conditions Used to Amplify Species Specific, Virulence Markers, and Antibiotic Resistance Genes in P. aeruginosa Isolates by PCR
| Virulence Factor | Target Gene | Primer Name | Sequence (5’ to 3’) | Length, bp | Annealing Temperature, °C | Amplicon Size, pb | References |
|---|---|---|---|---|---|---|---|
| Exotoxin A | exoA | ExoA-F | CCTCAGCATCACCAGGGA | 18 | 58 | 273 | This study |
| ExoA-R | CTGACGAAGAAGGTGGCATC | 20 | |||||
| Phosphplipase C | plcH | plc H-F | CCGCAACGACAAGTGGATG | 19 | 60 | 323 | This study |
| Plc H-R | AGCCGCCTTCCTGGTAGAC | 19 | |||||
| Elastase | lasB | las B-F | AGCCATCACCGAAGTCAAGG | 20 | 60 | 251 | This study |
| las B-R | CGGGAATCAGGTAGGAGACG | 20 | |||||
| Alginate | algD | Alg-D-F | TGGCGTTCGGACTTCTCG | 18 | 58 | 333 | This study |
| Alg-D-R | TGGTTTGGGCTATGTCGGTG | 20 | |||||
| beta-lactamase IMP | bla-IMP | Bla-IMP-F | TTGGTGGTTCTTGTAAATGCTG | 22 | 58 | 241 | This study |
| Bla-IMP-R | TTGTGGCTTGAACCTTACCG | 20 |
3.8. Detection of Virulence Genes by PCR
The PCR method was carried out for detection and distribution of 4 virulence genes algD, lasB, plcH, and exoA among 80 P. aeruginosa hospital isolates, using specific primers listed in Table 1. Extraction, PCR reactions, and gel electrophoresis were performed, as described previously.
3.9. Detection of blaIMP Resistance Gene by PCR
The presence of IMP genes in the isolates was assessed by the PCR technique using specific sets of forward and reverse primers, as shown in Table 1. PCR reactions were performed, as described previously.
3.10. Statistical Analysis
All statistical analyzes of data were done by the SPSS software, version 16.0 (SPSS Inc, Chicago, IL, USA, 2014). The values are presented as percentages. Pearson’s Chi-square test was applied for analysis and comparison of the frequency. P values ≤ 0.05 were assumed statistically significant.
4. Results
In the current study, a total of 80 P. aeruginosa strains were investigated. All isolates were primary detected by biochemical tests and finally confirmed by the molecular method (exoA gene PCR test). Totally, 80 isolates were obtained from hospitalized patients. Table 2 displays the demographic data of studied patients. Pseudomonas aeruginosa spp. infected patients consisted of 49 males (61.25%) and 31 females (38.75%). Maximum number of cases were in the age group of less than 30 years (n = 39, 48.75%) (Table 2). Eleven (13.75 %) patients had higher than 60 year age. The maximum number of infected patients were hospitalized in the burn ward (n = 34, 42.5%).
A total of 42 (52.5%) isolates recovered from wound specimens. Table 3 shows the antibiotic susceptibility profile of P. aeruginosa studied isolates. According to the in vitro antibiotic susceptibility testing (AST), colistin was the most effective antibiotic against isolates (97.5% isolates were susceptible) and levofloxacin was the least effective drug (67.5% isolates were resistant) (Table 3). Susceptibility rates for imipenem, amikacin, ceftazidime and ciprofloxacin were 41.25%, 40%, 38.75%, and 35%, respectively, and the susceptibility rate for doripenem, gentamicin, cefepime, piperacillin-tazobactam, and carbenicillin was 42.5%, 38.75%, 36.25%, 51.25%, and 43.75%, respectively (Table 3). Overall, the resistant rate among strains isolated from the wound was more than the urine isolates.
Demographic Features of the Patients Infected with Pseudomonas aeruginosa (N = 80)
| Category | Results (%) |
|---|---|
| Gender | |
| Male | 49 (61.25) |
| Female | 31 (38.75) |
| Age, y | |
| < 30 | 39 (48.75) |
| 30 - 44 | 19 (23.75) |
| 45 - 59 | 11(13.75) |
| > 60 | 11 (13.75) |
| City | |
| Yasuj | 40 (50) |
| Shiraz | 40 (50) |
| Hospital wards | |
| Burn | 34 (42.5) |
| General internal medicine | 9 (11.25) |
| ICU | 8 (10.0) |
| Pediatric/neonatal | 18 (22.5) |
| Surgery | 3 (3.75) |
| Gynecology | 8 (10.0) |
| Specimen | |
| Wound | 42 (52.5) |
| Urine | 38 (47.5) |
Antibiotic Resistance Profile of Studied Pseudomonas aeruginosa Isolates According to Specimen Type
| Source | Number (%) of Resistant Isolates | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PRL | TC | PTZ | LEV | CIP | MEM | IMI | DOR | ATM | CAZ | CPM | GM | TN | AK | PB | PY | |
| Urine (n=38) | 18 (47.36) | 20 (52.63) | 13 (34.21) | 24 (63.15) | 21 (55.26) | 20 (52.63). | 18 (47.365) | 18 (47.36) | 19 (50) | 20 (52.63) | 22 (58.79) | 22 (57.89) | 19 (50) | 20 (52.63) | 1 (2.63) | 21 (55.26) |
| Wound (n=42) | 27 (64.28) | 31 (73.80) | 26 (61.9) | 30 (71.42) | 31 (73.80) | 30 (71.42) | 29 (69) | 28 (66.66) | 29 (69) | 29 (69) | 29 (69) | 27 (64.28) | 28 (66.66) | 28 (66.66) | 1 (2.38) | 32 (76.19) |
| Total (n=80) | 45 (56.25) | 51 (63.75) | 39 (48.75) | 54 (67.5) | 52 (65) | 50 (62.5) | 47 (58.75) | 46 (57.5) | 48 (60) | 49 (61.25) | 51 (63.75) | 49 (61.25) | 47 (58.75) | 48 (60) | 2 (2.5) | 53 (66.25) |
Table 4 depicts the multiple antibiotic resistance patterns in isolates. The most common patterns were CIPR-CPMR-GEMR (47 isolates), which were followed by CIPR-CAZR-TOBR (46 isolates) and GEMR-TOBR-AKR (46 isolates). Analysis of cross-resistance results revealed that more than 52.5% of the isolates were resistant to carbapenems (imipenem, meropenem, doripenem) and 42 (52.5%) to imipenem, meropenem, ceftazidime, and cefepime (Table 4). A total of 55% of studied isolates were resistant to tobramycin, meropenem, levofloxacin, and ceftazidime. Based on the in vitro antibiotic susceptibility assay using disk diffusion technique, totally, 47 (58.75%) isolates were identified as MDR (32 isolates in Yasuj vs 15 isolates in Shiraz) (Table 5). A total of 24 (51%) MDR infected patients were hospitalized in the burn ward, and 57.44% MDR isolates were recovered from wound specimens. Only 1 (2.12%) MDR strain was resistant to colistin. Totally, 24 (51.06%) MDR strains were MHT-positive and 100% MDR isolates carried the lasB and plcH gene, whereas respectively, 86.66% and 100% of the isolates in Shiraz and Yasuj carried the algD gene. In this study 42 (52.5%) isolates were resistant to all tested carbapenems (imipenem, meropenem, doripenem) (Table 4). On the other hand, 5 isolates, which showed multi-drug resistance in disk diffusion test, were carbapenem susceptible.
Antibiotic Co-Resistance Patterns of Pseudomonas aeruginosa Isolated from Hospitalized Patients (n = 80)
| Pattern | Antibiotic Resistance Patterns | No. (%) |
|---|---|---|
| A | IMI, MEM, CPM, CAZ | 42 (52.5) |
| B | MEM, LEV, CAZ, TOB | 44 (55) |
| C | IMI, AK, CAZ, CIP, PB | 1 (1.25) |
| D | CIP, CPM, GEM | 47 (58.75) |
| E | CIP, CAZ, TOB | 46 (57.5) |
| F | CAZ, AK, PTZ | 42 (52.5) |
| G | GEM, TOB, AK | 46 (57.5) |
| H | IMI, MEM, DOR | 42 (52.5) |
| I | AK, PTZ, PB | 1 (1.25) |
| J | PRL, TC, AK | 44 (55) |
| K | PTZ, AK, CAZ | 42 (52.5) |
| L | AK, IMI, CAZ | 44 (55) |
Comparison of Carbapenem Resistant, MDR+, MHT+ Strains, and Virulence and Antibiotic Resistance Genes Among Pseudomonas aeruginosa Isolates According to Specimen, City, Hospital Ward in Shiraz and Yasuj, South-West of Iran (n = 80)
| Number of Isolates (%) | Carbapenem Resistant (n=42, 52.5%) | MDR+ (n=47, 58.75%) | MHT+ (n=24, 30%) | MDR+, MHT+ (n=24, 30%) | bla IMP+ (n=39, 48.75%) | MHT+, bla IMP+ (n=24, 30%) | plcH+ (n=80, 100%) | algD+ (n=78, 97.50%) | lasB+ (n=80, 100%) | lasB+, plcH+, algD+ (n=78, 97.50%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Specimen | |||||||||||
| Urine | 38 (47.5) | 18 (42.85) | 20 (42.55) | 10 (41.66) | 10 (41.66) | 18 (46.15) | 10 (41.66) | 38 (47.5) | 38 (48.72) | 38 (47.5) | 38 (48.72) |
| Wound | 42 (52.5) | 24 (57.15) | 27 (57.45) | 14 (58.34) | 14 (58.34) | 21 (53.85) | 14 (58.34) | 42 (52.5) | 40 (51.28) | 42 (52.5) | 40 (51.28) |
| Total | 80 (100) | 42 (100) | 47 (100) | 24 (100) | 24 (100) | 39 (100) | 24 (100) | 80 (100) | 78 (100) | 80 (100) | 78 (100) |
| City | |||||||||||
| Shiraz | 40 (50) | 14 (33.34) | 15 (31.92) | 11 (68.75) | 11 (45.83) | 15 (38.46) | 11 (45.84) | 40 (50) | 38 (48.72) | 40 (50) | 38 (48.72) |
| Yasuj | 40 (50) | 28 (66.67) | 32 (68.08) | 13 (43.35) | 13 (54.16) | 24 (61.54) | 13 (54.16) | 40 (50) | 40 (51.28) | 40 (50) | 40 (51.28) |
| Total | 80 (100) | 42 (100) | 47 (100) | 24 (100) | 24 (100) | 39 (100) | 24 (100) | 80 (100) | 78 (100) | 80 (100) | 78 (100) |
| Hospital wards | |||||||||||
| Burn | 34 (42.5) | 23 (54.76) | 24 (51.06) | 12 (48) | 12 (50) | 19 (48.71) | 12 (50) | 34 (42.5) | 32 (41.02) | 34 (42.5) | 32 (41.02) |
| Pediatric / neonatal | 18 (22.5) | 10 (23.81) | 11 (23.4) | 5 (35.71) | 5 (20.83) | 10 (25.64) | 5 (20.83) | 18 (22.5) | 18 (23.07) | 18 (22.5) | 18 (23.07) |
| Gynecology | 8 (10) | 3 (7.14) | 4 (8.51) | 2 (66.66) | 2 (8.33) | 3 (7.69) | 2 (8.334) | 8 (10) | 8 (10.25) | 8 (10) | 8 (10.25) |
| General internal medicine | 9 (11.25) | 4 (9.52) | 6 (12.7) | 3 (75) | 3 (12.5) | 4 (10.26) | 3 (12.5) | 9 (11.25) | 9 (11.54) | 9 (11.25) | 9 (11.54) |
| Surgery | 3 (3.75) | 1 (2.38) | 1 (2.13) | 1 (100) | 1 (4.17) | 1 (2.56) | 1 (4.16) | 3 (3.75) | 3 (3.85) | 3 (3.75) | 3 (3.85) |
| ICU | 8 (10) | 1 (2.38) | 1 (2.13) | 1 (100) | 1 (4.17) | 2 (5.13) | 1 (4.16) | 8 (10) | 8 (10.25) | 8 (10) | 8 (10.25) |
| Total | 80 (100) | 42 (100) | 47 (100) | 24 (100) | 24 (100) | 39 (100) | 24 (100) | 80 (100) | 78 (100) | 80 (100) | 78 (100) |
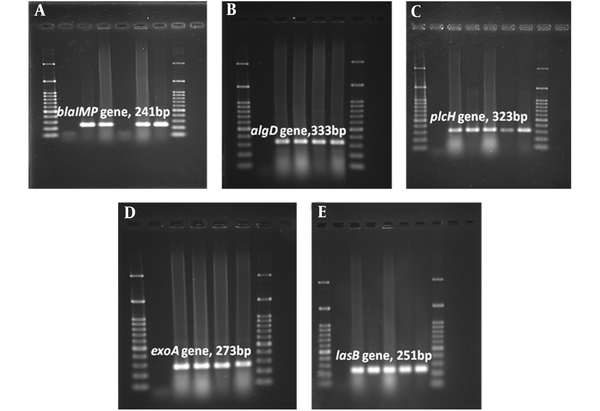
According to MHT results, 30% of isolates were carbapenemase producer (MHT+). A total of 50% of MHT-positive strains were recovered from patients hospitalized in burn ward. The prevalence rate of wound isolated MHT+ strains was higher in Yasuj hospitals (n = 9, 30%) than Shiraz hospitals (n = 5, 31.25%). All MHT+ isolates carried the blaIMP gene and also algD, lasB, and plcH virulence genes. Among 47 isolates which showed resistance to imipenem in disk diffusion test, 39 (82.97%) isolates carried blaIMP resistance gene in the PCR assay. The results of the PCR assay showed 93.61% of imipenem resistant isolates carried all studied virulence genes (Table 5). Images of gel electrophoresis used for detection of the amplified virulence factors and resistance genes in P. aeruginosa isolates was shown in Figure 1. The lasB and plcH were the most common virulence genes in P. aeruginosa isolates; the prevalence of these genes was 100%. In addition, algD virulence gene was detected in strains isolated from a wound with the frequency of 95.24% (n = 40). In this study, algD gene was the least virulence gene detected among all isolates in Shiraz (95%; n = 38). In MHT positive isolates, all surveyed virulence markers (plcH (100%), lasB (100%), and algD (100%) were detected (Table 5).
Agarose Gel Electrophoresis of the Amplified Resistance (blaIMP) and Virulenc (algD, plcH, exoA, lasB) Genes by PCR Assay. A, blaIMP gene (241 bp); B, algD gene (333 bp); C, plcH gene (323 bp); D, exoA gene (273 bp); E, lasB gene (251 bp). 100 bp size marker (DNA ladder) was used in agarose gel electrophoresis.
Among studied isolates, plcH+, and lasB+ genotype (100% isolates) were the most common virulence gene patterns; which were followed by algD+, plcH+, and lasB+ genotype (97.5% isolates) (Table 6). Out of the 80 P. aeruginosa isolates tested, 39 (48.75%) showed the algD+, plcH+, lasB+, and IMP+ genotype (Table 6). Figure 1 showed PCR results for the detection of virulence factors and resistance gene in P. aeruginosa isolates. Comparison of resistance profile, virulence, and antibiotic resistance genes among isolates collected from different specimens in 2 hospitals in Yasuj and Shiraz are summarized in Table 7. Among studied strains 39 isolates carried the blaIMP gene. In the present study, the highest prevalence of blaIMP resistance gene was observed in strains isolated from wound specimens with a frequency of 50% (n = 21). The blaIMP resistance gene was detected in all MHT positive and MDR isolates. All blaIMP gene positive strains carried all studied virulence genes (lasB, plcH and algD).
Virulence and Resistance Genes Patterns in Pseudomonas aeruginosa Isolates
| Pattern | Virulence and Resistance Genes | Total 80 (100%) |
|---|---|---|
| A | algD+, plcH+, lasB+, IMP+ | 39 (48.75) |
| B | algD+ , plcH+, lasB+ | 78 (97.5) |
| C | plcH+, lasB+, IMP+ | 39 (48.75) |
| D | algD+, lasB+, IMP+ | 39 (48.75) |
| E | plcH+, lasB+ | 80 (100) |
| F | algD+, plcH+ | 78 (97.5) |
| G | algD+, lasB+ | 78 (97.5) |
| H | lasB+, IMP+ | 39 (48.75) |
| K | plcH+, IMP+ | 39 (48.75) |
| L | algD+, IMP+ | 39 (48.75) |
Comparison of Resistance Profile, Virulence, and Antibiotic Resistance Genes Among Pseudomonas aeruginosa Isolates Collected from Different Specimens in Yasuj and Shiraz, South-West of Iran (n = 80)
| City | Total (n = 80, 100%) | ||||
|---|---|---|---|---|---|
| Shiraz (n = 40) | Yasuj (n = 40) | ||||
| Wound (n = 20, 50%) | Urine (n = 20, 50%) | Wound (n = 22, 55%) | Urine (n = 18, 45%) | ||
| Hospital Ward | |||||
| Burn | 8 (40) | 5 (25) | 17 (77.27) | 4 (22.22) | 34 (42.5) |
| General internal medicine | 0 (0) | 6 (30) | 1 (4.54) | 2 (11.11) | 9 (11.25) |
| ICU | 2 (10) | 6 (30) | 0 (0) | 0 (0) | 8 (10) |
| Pediatric | 4 (20) | 1 (5) | 2 (9.09) | 10 (55.55) | 17 (21.25) |
| Surgical | 2 (10) | 1 (5) | 0 (0) | 0 (0) | 3 (3.75) |
| Gynecology | 4 (20) | 1 (5) | 2 (9.09) | 2 (11.11) | 9 (11.25) |
| Antibiotic Resistance Profile | |||||
| IMI | 11 (55) | 6 (30) | 18 (81.81) | 12 (66.66) | 47 (58.75) |
| GM | 8 (40) | 8 (40) | 19 (86.36) | 14 (77.77) | 49 (61.25) |
| CAZ | 8 (40) | 6 (30) | 21 (95.45) | 14 (77.77) | 49 (61.25) |
| CO | 0 (0) | 1 (5) | 1 (4.54) | 0 (0) | 2 (2.5) |
| CIP | 11 (55) | 8 (40) | 20 (90.90) | 13 (72.22) | 52 (65) |
| AK | 8 (40) | 6 (30) | 20 (90.90) | 14 (77.77) | 48 (60) |
| CPM | 8 (40) | 8 (40) | 21 (95.45) | 14 (77.77) | 51 (63.75) |
| PTZ | 7 (35) | 6 (30) | 19 (86.36) | 7 (38.88) | 39 (48.75) |
| TIC | 11 (55) | 7 (35) | 20 (90.90) | 13 (72.22) | 51 (63.75) |
| MHT(+) strains | 5 (31.25) | 6 (37.5) | 9 (30) | 4 (13.33) | 24 (52.17) |
| MDR (+) strains | 8 (40) | 7 (35) | 19 (86.36) | 13 (72.22) | 47 (58.75) |
| blaIMP + strains | 8 (40) | 7 (35) | 13 (59.09) | 11 (61.11) | 39 (48.75) |
| Virulence Markers | |||||
| algD+ | 18 (90) | 20 (100) | 22 (100) | 18 (100) | 78 (97.5) |
| lasB+ | 20 (100) | 20 (100) | 22 (100) | 18 (100) | 80 (100) |
| plcH+ | 20 (100) | 20 (100) | 22 (100) | 18 (100) | 80 (100) |
5. Discussion
In the present cross-sectional study, we conducted a multicenter study to assess the prevalence of antibiotic resistance profiles, multidrug resistance, carbapenemase production, and virulence determinants in P. aeruginosa isolated from clinical specimens in South-west Iran (cities of Yasuj and Shiraz). According to the exoA gene PCR results, all 80 Pseudomonas isolates were identified as P. aeruginosa. Most of the P. aeruginosa isolates were recovered from burn patients (n = 34; 42.50%), which were followed by paediatrics (n = 18; 22.5%). The prevalence rate of wound isolated strains was higher in Yasuj hospitals (55%) than Shiraz hospitals (50%). The observations on the prevalence of P. aeruginosa strains in clinical specimens were similar to those of other authors noting that the isolation rate of this bacterium from wounds was more than that of urine among clinical specimens.
Antibiotic resistance among P. aeruginosa is a global problem. The presence of multiple potential virulence factors and resistance to clinically relevant antibiotics, especially emergence of MDR strains, and their ability to transfer resistance genes to other bacteria such as Enterobacteriaceae family and other Gram negative non-fermenter bacteria (e.g., Acinetobacter spp. and Stenotrophomonas maltophilia), have made them a subject of close scrutiny and in-depth investigations (22, 23). Carbapenems, ceftazidime, quinolones, and aminoglycosides are important drugs for the treatment of Pseudomonas infections. It is important to highlight the presence of the 56 imipenem-resistant, 54 ceftazidime-resistant, 55 ciprofloxacin-resistant, and 52 amikacin resistant P. aeruginosa isolates in this study. Similar results have been reported by Yayan et al. (24). Recently, it has been relatively common to find resistant to carbapenems, as a result of carbapenemase production and modifications in Por proteins (23, 25). Long-term use of these antimicrobials in hospitals, community, and veterinary practice in Iran may be the primary reason for the presence of beta-lactam and carbapenem resistant isolates. Furthermore, cefepime, ciprofloxacin, and carbenicillin resistance (63.75%, 65%, and 66.25%, respectively) in this study was quite high in accordance with findings reported by Burgess et al. (26).
Depending on the MHT results, the current study revealed that the P. aeroginosa isolates showed a high rate of carbapenemase production (30% isolates were MHT+); this was in accordance with the findings of other studies, showing the high prevalence of resistanceto carbapenem among clinical P. aeroginosa strains (9, 25). In present research, 70% studied strains were found to be resistant to imipenem, and strains showing the same resistance to meropenem, however, the rate of resistance to doripenem was lower (63.75%). In this study, the prevalence of carbapenem resistant was higher than that of another report on the prevalence of imipenem resistant P. aeruginosa (IRPA) (58.7%) in Ahvaz, Iran (27). A similar enhancement in the prevalence of IRPA has been noticed earlier in several studies reported from different countries (28). Of all 56 imipenem resistant P. aeruginosa (IRPA) isolates, 39 (69.64%) carried the bla-IMP gene, while this gene was detected in all of MHT+ isolats, as reported by another study carried out in Tehran, Iran. In our study there was a significant difference between bla-IMP gene positive and MHT positive isolates (P = 0.0001), and also between antibiotic resistance and existence of the bla-IMP gene (P = 0.0001).
The progressive increase in carbapenem resistance prevalence in Iran is alarming; therefore, carbapenems cannot serve as the drug of choice for treating multidrug resistant P. aeruginosa infections in the near future. It should also be noted that in many cases antibiotic resistance is transmitted to humans and hospital environment through other sources. As reported, the selection of antibiotics for the treatment of P. aeruginosa infections was very limited and isolates were resistant to most of the common drugs. Resistant strains were tested for alternative therapeutic options such as colistin that showed good response to this drug (97.5% isolates were sensitive). According to the obtained results, the rate of resistance among carbapenem resistant (CR) strains to amikacin, ciprofloxacin, and ceftazidime was 100%. This was in contrast with a previous study from the Shiraz (Iran), which reported that ceftazidime is the most effective drug against this isolate (9). Due to many carbapenem-resistant (CR) and MDR P. aeruginosa isolates are susceptible to very few antibiotics other than colistin, treatment should involve a combination therapy including colistin.
In the present study, 2 isolates were resistant to colistin. This result is in contrast with a previous research from the South-west of Iran (Shiraz) and Germany, which reported all isolates were susceptible to colistin (2, 24). Colistin resistant isolates found in our study is quite alarming. Colistin stands as an antibiotic of last resort for multidrug-resistant infections. The potential risk for colistin resistance emerging in P. aeruginosa has important clinical implications in infection treatment protocols. The resistant to carbapenems in P. aeroginosa is usually encoded by blaIMP gene. The prevalence rate of blaIMP gene positive strains was higher in Yasuj hospitals (61.54%) than Shiraz hospitals (38.46%); however, this difference was statistically significant (P = 0.04). Another disturbing finding in our study was the relatively high prevalence of doripenem resistant P. aeroginosa (DRPA). The occurrence of DRPA among the P. aeruginosa isolates in our study was seen to be 63.75%. Furthermore, the PCR results of isolates confirmed that 34 DRPA isolates contained the blaIMP gene. These results were dissimilar to those of another report from France, showing the prevalence rate of DRPA to be 15% (29). In Iran research on DRPA isolates has not be performed and reports are not available. Despite the global spread of resistance to meropenem, as the most active antipseudomonal agent available, no potent and effective antipseudomonal antibiotics have been introduced during the past decade. Doripenem is a new parental carbapenem that is now being developed for intravenous use, however, resistance to this drug has been increased.
The presence of virulence determinants associated with P. aeruginosa enhanced their pathogenicity. Moreover, it was found that wound isolated strains had a larger number of virulence determinants, in comparison to urine isolated strains, as shown in a previous study. The present study revealed the higher frequency of the lasB, plcH, and algD virulence genes among P. aeruginosa isolates, while the frequency of the algD gene was not the same among wound isolates in Shiraz and Yasuj. Our results were similar to those of another study from Iran and other parts of the world, reporting the high prevalence of these virulence genes among P. aeruginosa and the high prevalence of the blaIMP gene among isolates (15). The exoA gene was detected in all of the 80 P. aeruginosa isolates, which was in agreement with the results reported by other investigators testing tested P. aeruginosa strains for the presence of the exoA gene (30, 31).
Our results showed that the prevalence rate of MDR strains is higher in Yasuj isolates (68.08%) than Shiraz isolates (31.92%); however, this difference was statistically significant (P = 0.0001). Overall, in the present study, the distribution of resistance gene was more common in multidrug resistant isolates than in non-MDR isolates and the high prevalence of multiple pathogenesis determinants could potentially contribute to bacterial colonization and pathogenesis of P. aeruginosa in the human community. The higher prevalence of blaIMP gene in MDR+ and MHT+ isolates, may explain the role of this gene in emergence of resistance to the carbapenem antibiotics. The algD gene was detected in 97.5% of all tested isolates, with burn ward isolates (n = 32, 41.02%) showing higher prevalence than other hospital wards isolates [paediatric/neonatal (23.07), general internal medicine (11.54%), ICU (10.25%), gynecology (10.25%), and surgery (3.85%)]. This was in accordance with the findings of another study identifying the algD gene in about 91.1% of P. aeruginosa strains (15). To conclude, algD gene was found to be more common among MDR isolates than non-MDR isolates; also, it had a considerable ability to show other virulence genes and drug resistance.
Among studied strains 39 (48.75%) isolates carried the blaIMP gene, and 24 (30%) isolates were identified as MHT+, nevertheless, this difference was statistically significant (P = 0.0001). Among MHT+ isolates, only 15 isolates carried the blaIMP gene. Likewise, all MHT+ isolates were multi-drug resistant (MDR). This results show that the some of the strains that carry the blaIMP gene may not be detectable by MHT phenotypic test or other genes and mechanisms involved in the resistant to carbapenems. Emergence of multi-drug resistant P. aeruginosa, especially to carbapenems and aminoglycosides, and the high prevalence of virulence traits in our study could be regarded as an alarming situation (32, 33). Antibiotic resistant pathogenic bacteria can come from the variety of foods (34). Another interesting finding in our study was the more frequent occurrence of algD+, MDR, MHT+, and blaIMP+ strains among the P. aeruginosa isolates from Yasuj compared with those from Shiraz. Also, colistin resistant and pan-drug resistant P. aeruginosa (PDR-PA) strains are emerging. However, the major differences in the occurrence of resistance between the 2 regions were most likely due to the differences in the usage of therapeutic drugs in hospitals.
6. Conclusions
In our study the emergence and distribution of potentially highly pathogenic and carbapenem resistant strains in combination with a multidrug resistant pattern is alarming, as a possible outcome would be a severe clinical result concomitant with great limitations in antibiotic therapy. However, a regular surveillance of virulence determinants, monitoring of antibiotic susceptibility pattern, and designing practical guide antibiotic stewardship for P. aeruginosa are suggested.
Acknowledgements
References
-
1.
Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev. 2009;22(4):582-610. [PubMed ID: 19822890]. https://doi.org/10.1128/CMR.00040-09.
-
2.
Poorabbas B, Mardaneh J, Rezaei Z, Kalani M, Pouladfar G, Alami MH, et al. Nosocomial Infections: Multicenter surveillance of antimicrobial resistance profile of Staphylococcus aureus and Gram negative rods isolated from blood and other sterile body fluids in Iran. Iran J Microbiol. 2015;7(3):127-35. [PubMed ID: 26668699].
-
3.
Tiwari N, Rajdev S, Mullan S. Resistance Trends among Pseudomonas aeruginosa Isolates in a Tertiary Care Centre in South Gujarat. Adv Microbiol. 2017;7(3):188-94. https://doi.org/10.4236/aim.2017.73015.
-
4.
Lister PD. Chromosomally-encoded resistance mechanisms of Pseudomonas aeruginosa: therapeutic implications. Am J Pharmacogenomics. 2002;2(4):235-43. [PubMed ID: 12421094].
-
5.
Sadikot RT, Blackwell TS, Christman JW, Prince AS. Pathogen-host interactions in Pseudomonas aeruginosa pneumonia. Am J Respir Crit Care Med. 2005;171(11):1209-23. [PubMed ID: 15695491]. https://doi.org/10.1164/rccm.200408-1044SO.
-
6.
Lavoie EG, Wangdi T, Kazmierczak BI. Innate immune responses to Pseudomonas aeruginosa infection. Microbes Infect. 2011;13(14-15):1133-45. [PubMed ID: 21839853]. https://doi.org/10.1016/j.micinf.2011.07.011.
-
7.
Branski LK, Al-Mousawi A, Rivero H, Jeschke MG, Sanford AP, Herndon DN. Emerging infections in burns. Surg Infect. 2009;10(5):389-97. [PubMed ID: 19810827]. [PubMed Central ID: PMC2956561]. https://doi.org/10.1089/sur.2009.024.
-
8.
Rossolini GM, Mantengoli E. Treatment and control of severe infections caused by multiresistant Pseudomonas aeruginosa. Clin Microbiol Infect. 2005;11 Suppl 4:17-32. [PubMed ID: 15953020]. https://doi.org/10.1111/j.1469-0691.2005.01161.x.
-
9.
Anvarinejad M, Japoni A, Rafaatpour N, Mardaneh J, Abbasi P, Amin Shahidi M, et al. Burn Patients wounds Infected With Metallo-Beta-Lactamase-Producing Pseudomonas aeruginosa: Multidrug Resistant Strains. Arch Trauma Res. 2014;3(2). https://doi.org/10.5812/atr.18182.
-
10.
Meletis G, Exindari M, Vavatsi N, Sofianou D, Diza E. Mechanisms responsible for the emergence of carbapenem resistance in Pseudomonas aeruginosa. Hippokratia. 2012;16(4):303-7. [PubMed ID: 23935307].
-
11.
Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40(4):277-83. [PubMed ID: 25859123].
-
12.
Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis. 2011;17(10):1791-8. [PubMed ID: 22000347]. https://doi.org/10.3201/eid1710.110655.
-
13.
Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440-58. table of contents. [PubMed ID: 17630334]. https://doi.org/10.1128/CMR.00001-07.
-
14.
Zmarlicka MT, Nailor MD, Nicolau DP. Impact of the New Delhi metallo-beta-lactamase on beta-lactam antibiotics. Infect Drug Resist. 2015;8:297-309. [PubMed ID: 26345624]. https://doi.org/10.2147/IDR.S39186.
-
15.
Mitov I, Strateva T, Markova B. Prevalence of virulence genes among bulgarian nosocomial and cystic fibrosis isolates of pseudomonas aeruginosa. Braz J Microbiol. 2010;41(3):588-95. [PubMed ID: 24031533]. https://doi.org/10.1590/S1517-83822010000300008.
-
16.
Kipnis E, Sawa T, Wiener-Kronish J. Targeting mechanisms of Pseudomonas aeruginosa pathogenesis. Med Mal Infect. 2006;36(2):78-91. [PubMed ID: 16427231]. https://doi.org/10.1016/j.medmal.2005.10.007.
-
17.
Stehling EG, Silveira WD, Leite Dda S. Study of biological characteristics of Pseudomonas aeruginosa strains isolated from patients with cystic fibrosis and from patients with extra-pulmonary infections. Braz J Infect Dis. 2008;12(1):86-8. [PubMed ID: 18553021].
-
18.
Sonbol FI, Khalil MA, Mohamed AB, Ali SS. Correlation between antibiotic resistance and virulence of Pseudomonas aeruginosa clinical isolates. Turk J Med Sci. 2015;45(3):568-77. [PubMed ID: 26281322].
-
19.
Fazeli N, Momtaz H. Virulence Gene Profiles of Multidrug-Resistant Pseudomonas aeruginosa Isolated From Iranian Hospital Infections. Iran Red Crescent Med J. 2014;16(10). e15722. [PubMed ID: 25763199]. https://doi.org/10.5812/ircmj.15722.
-
20.
Lee VT, Smith RS, Tummler B, Lory S. Activities of Pseudomonas aeruginosa effectors secreted by the Type III secretion system in vitro and during infection. Infect Immun. 2005;73(3):1695-705. [PubMed ID: 15731070]. https://doi.org/10.1128/IAI.73.3.1695-1705.2005.
-
21.
Wayne PA; Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing. 2007.
-
22.
Beceiro A, Tomas M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev. 2013;26(2):185-230. [PubMed ID: 23554414]. https://doi.org/10.1128/CMR.00059-12.
-
23.
Mellouk FZ, Bakour S, Meradji S, Al-Bayssari C, Bentakouk MC, Zouyed F, et al. First Detection of VIM-4-Producing Pseudomonas aeruginosa and OXA-48-Producing Klebsiella pneumoniae in Northeastern (Annaba, Skikda) Algeria. Microb Drug Resist. 2017;23(3):335-44. [PubMed ID: 27314660]. https://doi.org/10.1089/mdr.2016.0032.
-
24.
Webber MA, Yayan J, Ghebremedhin B, Rasche K. Antibiotic Resistance of Pseudomonas aeruginosa in Pneumonia at a Single University Hospital Center in Germany over a 10-Year Period. Plos One. 2015;10(10). e0139836. https://doi.org/10.1371/journal.pone.0139836.
-
25.
Malkocoglu G, Aktas E, Bayraktar B, Otlu B, Bulut ME. VIM-1, VIM-2, and GES-5 Carbapenemases Among Pseudomonas aeruginosa Isolates at a Tertiary Hospital in Istanbul, Turkey. Microb Drug Resist. 2017;23(3):328-34. [PubMed ID: 27326514]. https://doi.org/10.1089/mdr.2016.0012.
-
26.
Burgess DS, Nathisuwan S. Cefepime, piperacillin/tazobactam, gentamicin, ciprofloxacin, and levofloxacin alone and in combination against Pseudomonas aeruginosa. Diagnos Microbiol Infect Dis. 2002;44(1):35-41. https://doi.org/10.1016/S0732-8893(02)00420-0.
-
27.
Farajzadeh Sheikh A, Rostami S, Jolodar A, Tabatabaiefar MA, Khorvash F, Saki A, et al. Detection of Metallo-Beta Lactamases Among Carbapenem-Resistant Pseudomonas aeruginosa. Jundishapur J Microbiol. 2014;7(8). https://doi.org/10.5812/jjm.12289.
-
28.
Onguru P, Erbay A, Bodur H, Baran G, Akinci E, Balaban N, et al. Imipenem-resistant Pseudomonas aeruginosa: risk factors for nosocomial infections. J Korean Med Sci. 2008;23(6):982-7. [PubMed ID: 19119440]. https://doi.org/10.3346/jkms.2008.23.6.982.
-
29.
Luyt CE, Aubry A, Lu Q, Micaelo M, Brechot N, Brossier F, et al. Imipenem, meropenem, or doripenem to treat patients with Pseudomonas aeruginosa ventilator-associated pneumonia. Antimicrob Agents Chemother. 2014;58(3):1372-80. [PubMed ID: 24342638]. https://doi.org/10.1128/AAC.02109-13.
-
30.
Dong D, Zou D, Liu H, Yang Z, Huang S, Liu N, et al. Rapid detection of Pseudomonas aeruginosa targeting the toxA gene in intensive care unit patients from Beijing, China. Front Microbiol. 2015;6:1100. [PubMed ID: 26500639]. https://doi.org/10.3389/fmicb.2015.01100.
-
31.
Mohammadzadeh A, Mardaneh J, Ahmadi R, Adabi J. Evaluation of the Virulence Features and Antibiotic Resistance Patterns of Pathogenic Pseudomonas aeruginosa Strains Isolated from Hospitalized Patients in Gonabad, Iran. Arch Pediatr Infect Dis. 2017;In press(In press). https://doi.org/10.5812/pedinfect.41267.
-
32.
Izadpour F, Ranjbari N, Aramesh MR, Moosavian M, ShahAli S, Larki F, et al. An Investigation of Antibacterial Resistance Patterns Among Acinetobacter baumannii and Pseudomonas aeruginosa Isolates Collected from Intensive Care Units of a University-Affiliated Hospital in Ahvaz, Iran. Jundishapur J Microbiol. 2016;9(8). https://doi.org/10.5812/jjm.35624.
-
33.
Aghazadeh M, Zahedi Bialvaei A, Aghazadeh M, Kabiri F, Saliani N, Yousefi M, et al. Survey of the Antibiofilm and Antimicrobial Effects of Zingiber officinale (in Vitro Study). Jundishapur J Microbiol. 2016;9(2). e30167. [PubMed ID: 27127591]. https://doi.org/10.5812/jjm.30167.
-
34.
Anvarinejad M, Pouladfar GR, Pourabbas B, Amin Shahidi M, Rafaatpour N, Dehyadegari MA, et al. Detection of Salmonella spp. with the BACTEC 9240 Automated Blood Culture System in 2008 - 2014 in Southern Iran (Shiraz): Biogrouping, MIC, and Antimicrobial Susceptibility Profiles of Isolates. Jundishapur J Microbiol. 2016;9(4). https://doi.org/10.5812/jjm.26505.