Abstract
Background:
Acinetobacter baumannii has appeared as an important opportunistic pathogen responsible for nosocomial infections. The rising trend of antibiotic resistance amongst A. baumannii isolates has become a global concern. The most prevalent procedure of resistance is beta-lactamase and carbapenemases production with genes on mobile elements.Objectives:
The aim of the current research was to assess antibiotic susceptibility schema and the frequency of TEM, PER, and NDM-1 genes among A. baumannii isolates.Methods:
One hundred and eighty three specimens from November 2014 to February 2015 were collected from Golestan and Imam Khomeini hospitals in Ahvaz, Iran. Drug susceptibility tests were carried out by Kirby-Bauer method. Extended spectrum-beta-lactamases (ESBLs) production was determined by the combination disk method and carbapenemases production was determined by the modified hodge test (MHT) according to the CLSI recommendations. TEM, PER, and NDM-1 were detected by PCR.Results:
Out of 183 Acinetobacter isolates, 151 (82.5 %) were identified as A. baumannii by standard chemical tests. The highest resistance was determined to ciprofloxacin (97.3 %), whereas the higher rate of susceptibility was observed to colistin (98.7%). 1.3% of the A. baumannii isolates were positive for ESBL in combined disc test. Production of carbapenemase was detected in 47.1% of the A. baumannii isolates using MHT. The prevalence of TEM and PER genes was 36.4 % and 25.1 %, respectively. NDM-1 genes were not detected.Conclusions:
The prevalence of carbapenemase positive A. baumannii isolates in the current study makes a serious concern and highlights the need for infection control through antibiotic management protocols and rapid detection of resistant strains.Keywords
Antibiotic Resistance ESBL Carbapenemase Acinetobacter baumannii
1. Background
Acinetobacter baumannii has appeared as an important bacterial pathogen associated with hospital acquired infections. Acinetobacter baumannii is often related to pneumonia, urinary tract, bloodstream, wound, and nosocomial meningitis infections (1). Acinetobacter baumannii strains have become a global concern because of their increasing rate of antibiotic resistance. This bacterium has tendency to gain resistance quickly to various types of antibiotics and thus, displays numerous rates of resistance to different antibacterial drugs (2). Gram-negative bacteria have evolved two major genetic pathways for resistance to beta-lactams including mutation of special intrinsic genes and acquisition of resistance genes.
Extended spectrum-beta-lactamases (ESBLs) genes represent the main mechanism of resistance to β-lactam antibiotics against later generation cephalosporins such as cefepime, cefotaxime, and ceftazidime in bacteria. These beta lactamase enzymes are often found in Enterobacteriaceae, Pseudomonas aeruginosa, and A. baumannii and are isolated from bacteria worldwide (3). Most of ESBLs are members of either TEM, SHV, CTX-M or PER (class A) families according to the Ambler molecular classification of β-lactamase genes (4). Carbapenems are the most effective antibiotics used to treat A. baumannii infections due to multidrug resistant A. baumannii strains. Whether resistance to carbapenem is related to an increased risk of mortality in patients with A. baumannii infections is controversial. The emergence of resistance to carbapenems among A. baumannii isolates in serious infections is now a significant threat to public health (5). Resistance to carbapenem is almost caused by the production of enzymes- carbapenemases that hydrolyze carbapenems and other β-lactams. Carbapenemase enzymes are divided into three Ambler classes of beta-lactamase A, B, and D (5).
Prevention and control of nosocomial infections caused by multidrug resistant A. baumannii strains require maintaining a clean healthcare environment, sufficient reprocessing of reusable medical devices, adequate capacity of healthcare systems for contact isolation, suitable capability of microbiological laboratories, and having a hospital hygiene team. Furthermore, awareness about resistant A. baumannii strains sources in healthy units improves the care of infection transmission and prevention from spreading of these infections (6, 7).
2. Objectives
The purpose of the current research was to find out extended spectrum B-lactamase and carbapenemases production by two different phenotypic methods and to look for bla TEM, bla PER, and bla NDM-1genes among A. baumannii clinical isolates in two teaching hospitals in Ahvaz.
3. Methods
3.1. Bacterial Isolates
A total of 183 non-duplicated clinical isolates of Acinetobacter were prepared between December 2014 and October 2015 from two different university-affiliated hospitals, Golestan and Imam Khomeini, in Ahvaz city, Iran. The isolates were recovered from different specimens, including tracheal aspirate, wound, blood, urine, abscess, pleural fluid, and CSF. Acinetobacter baumannii was the most prevalent species with 151 (82.5%) isolates. The rest (17.5%) were identified as Acinetobacter spp. Bacterial isolates were recognized by conventional biochemical tests including Gram staining, oxidase test, fermentative and oxidative properties, growth at 42°C, and citrate and malonate tests (8). In addition, A. baumannii species were confirmed for bla OXA-51-like genes by PCR (9).
3.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing and interpretation were employed for all isolates against 11 antimicrobial agents including imipenem (IPM: 10 µg), meropenem (MEM: 10 µg), ceftazidime (CAZ: 30 µg), cefotaxime (CTX: 30 µg), cefepime (FEP: 30 µg), ciprofloxacin (CIP: 5 µg), piperacillin (PIP, 100 µg), amikacin (AK: 30 µg), gentamicin (GEN: 30 µg), tetracycline (TE: 30 µg), and colistin (CO, 10 µg) (mast group, UK). Susceptibility test was carried out by Kirby-Bauer disk diffusion method on Mueller-Hinton agar (MHA) (Merck, Germany) according to the 2014 clinical laboratory standards institute (CLSI) guidelines (10). Susceptibility tests were monitored using Escherichia coli ATCC 25922 strain.
3.3. ESBL Phenotypic Detection of Isolates
Identification of ESBLs was performed for the isolates by combination disk test (CDT). The isolates were examined for the inhibition zone of ceftazidime (CAZ) 30 µg + clavulanic CA10 µg adjacent to disk containing ceftazidime (CAZ) alone and cefotaxime (CTX) 30 µg + clavulanic CA 10 µg adjacent to disk containing cefotaxime alone on Muller Hinton agar (Mast Group, UK). ESBL test was considered positive if the inhibition zone diameter in presence of clavulanic acid was ≥ 5 mm larger than that in the lack of it. Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC700603 were used as negative and positive controls for ESBL production, respectively (10).
3.4. Detection of Carbapenemases Producing Isolates
The carbapenem-resistant strains were evaluated for carbapenemase production by modified hodge test (MHT) according to the CLSI guidelines. Escherichia coli ATCC25922 (a sensitive indicator strain to carbapenems) was inoculated in sterile saline to achieve 0.5 McFarland turbidity standard and then was cultured onto Mueller-Hinton agar (MHA) (Merck, Germany) plate. After brief drying, ertapenem (10 µg) disc was placed at the center of the plate on the lawn culture. The test strains were heavily inoculated from the edge of the ertapenem disc to the periphery of the plate in three different directions. Three isolates were inoculated in one plate. The plates were incubated at 35°C overnight. The presence of distorted or a cloverleaf shaped zone of inhibition was considered as positive test for carbapenemase production strains according to the CLSI recommendations. Klebsiella pneumoniae ATCC BAA -1705 was used in this study for quality control (10).
3.5. Isolation of Genomic DNA
Total genomic DNAs of the bacterial isolates were prepared by using boiling method for DNA extraction. Briefly, at least 5 colonies were harvested from MacConkey agar plate and suspended in 500 µL TE buffer. The suspensions were incubated at 95°C for 15 minutes. Then, the cells were pelleted at the bottom of the tube by centrifugation at 12,000 × g for 5 minutes. The supernatants were transferred to new tubes and used as templates for PCR reaction.
3.6. Detection of bla OXA-51, bla TEM, bla PER and bla NDM-1 Genes by PCR
Clinical isolates of A. baumannii were confirmed by PCR assay with specific primer for presence of bla OXA-51-like gene listed in Table 2. Acinetobacter baumannii (ATCC 19606) was used as positive control (9). The ESBL genes determination was carried out by PCR to detect bla TEM and bla PER genes (11). Klebsiella pneumonia strain 7881 and Pseudomonas aeruginosa KOAS strain were used as positive control strains in the determination of these genes, respectively (11). Carbapenem-resistant strains were screened for bla NDM-1 gene by using specific primers targeting bla NDM-1. Klebsiella pneumonia NDM 2146 was used as control strain in the detection of these genes (12). Primer pairs used for the amplification of these genes are shown in Table 2.
Antibiotic Resistant Pattern of A. baumannii Isolates
| Antibiotics | Resistant (%) | Intermediate (%) | Sensitive (%) |
|---|---|---|---|
| Imipenem | 138 (91.4) | 2 (1.3) | 11 (7.3) |
| Meropenem | 142 (94) | 1 (0.7) | 8 (5.3) |
| Ceftazidime | 144 (95.4) | 0 (0) | 7 (4.6) |
| Cefotaxime | 141 (93.4) | 5 (3.3) | 5 (3.3) |
| Cefepime | 139 (92) | 2 (1.3) | 10 (6.7) |
| Ciprofloxacin | 147 (97.4) | 0 (0) | 4 (2.6) |
| Amikacin | 143 (94.7) | 0 (0) | 8 (5.3) |
| Piperacillin | 142 (94) | 0 (0) | 9 (6) |
| Gentamicin | 142 (94) | 0 (0) | 9 (6) |
| Tetracycline | 103 (68) | 16 (11) | 32 (21) |
| Colistin | 2 (1.3) | 0 (0) | 149 (98.7) |
The Primer Sequences Used in This Study
| Gene | Primer Sequences 5’ → 3’ | Product Size (bp) | Annealing (°C) | Reference |
|---|---|---|---|---|
| bla OXA-51 | F: TAATGCTTTGATCGGCCTTG | 353 | 54 | 7 |
| R: TGGATTGCACTTCATCTTGG | ||||
| bla TEM | F: GAGTATTCAACATTTCCGTGTC | 800 | 58 | 9 |
| R: TAATCAGTGAGGCACCTATCTC | ||||
| bla PER | F: AATTTGGGCTTAGGGCAGAA | 925 | 54 | 9 |
| R: ATGAATGTCATTATAAAAGC | ||||
| bla NDM-1 | F: GGTTTGGCGATCTGGTTTTC | 621 | 52 | 10 |
| R: CGGAATGGCTCATCACGATC |
To amplify these genes, each reaction was performed in a final volume of 25 µL containing 1x PCR buffer, 1U Taq polymerase, 1.5 mM MgCl2, 200 µM of dNTP, 10 pmol of each primer (Bioneer, South Korea), and 1 µL of extracted DNA. A negative control without DNA template was included in each PCR experiment. Amplification conditions were programmed in Master cycler Eppendorf (Eppendorf, Germany). Amplification conditions for PCR were as follows: Initial denaturation at 94°C for 3 minutes; 35 cycles of 94°C for 45 seconds, annealing at 53 - 58°C for 45 seconds, extension at 72°C for 1 minute, and final extension at 72°C for 5 minutes.
3.7. Electrophoresis
PCR products were electrophoresed on 1.5% agarose gel (Fermentase, Germany) and visualized under UV gel documentation system.
4. Results
In the present study, 183 non-repetitive isolates of Acinetobacter were recovered from hospitalized patients admitted to Golestan and Imam Khomeini teaching hospitals during December 2014 to October 2015 in Ahvaz city, Iran. 151 (82.5%) isolates were identified as A. baumannii by standard chemical tests. The remaining isolates (17.5%) were identified as Acinetobacter spp. Of the 151 A. baumannii isolates, 66 (43.7%) were obtained from female patients and 85 (56.3%) from males. Of the 151 isolates, 88 (58.3%) were isolated from tracheal aspirate, 26 (17.1%) from blood, 19 (12.6%) from wound, 5 (3.3%) from urine, 5 (3.3%) from CSF, 4 (2.7%) from abscess, and 4 (2.7%) from pleural fluid.
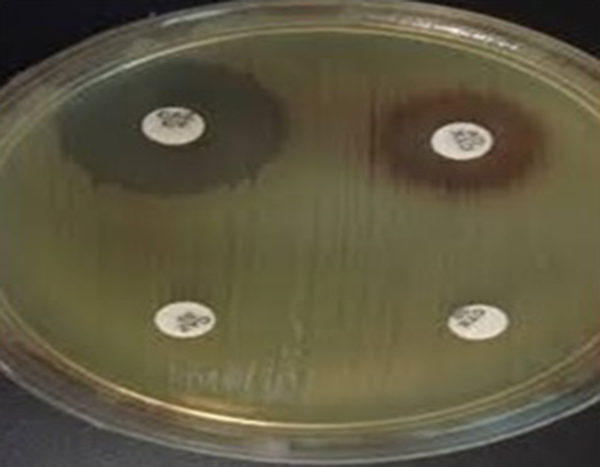
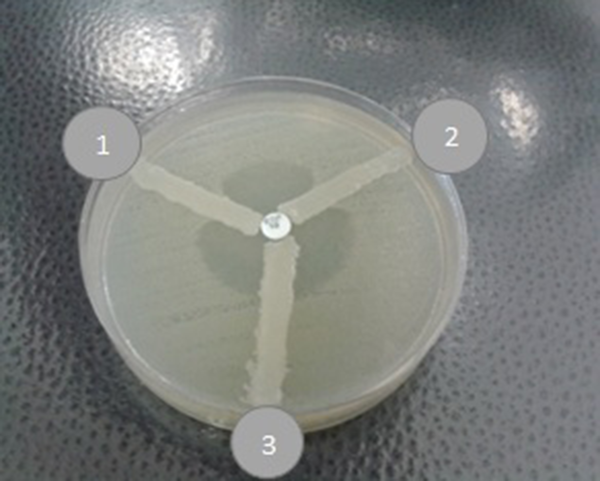
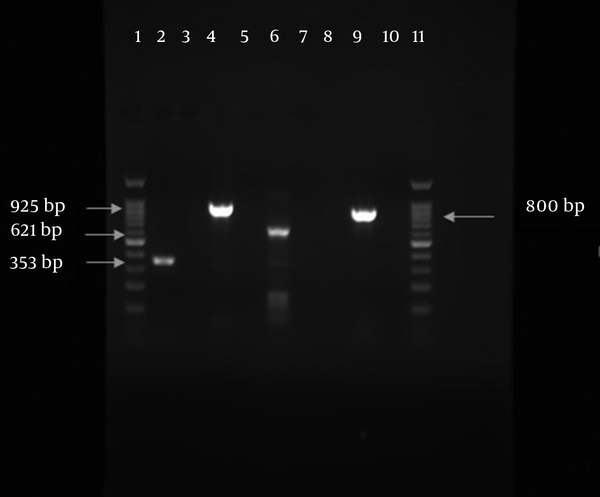
In the antimicrobial susceptibility test, A. baumannii isolates displayed resistance pattern as follows: 147 (97.4%) to ciprofloxacin, 144 (95.4%) to ceftazidime, 141 (93.4%) to cefotaxime, 139 (92%) to cefepime, 143 (94.7%) to amikacin, 142 (94%) to piperacillin, 142 (94%) to meropenem, 138 (91.4%) to imipenem, 142 (94%) to gentamicin, 103 (68%) to tetracycline, and 2 (1.3%) to colistin (Table 1). A high susceptibility rate was observed to colistin. All the ceftazidime resistant isolates were tested by combined disk test for assessing ESBL production (Figure 1). Among 144 ceftazidime resistant A. baumannii strains, only 2 isolates (1.3%) were positive for ESBL. Modified Hodge test was performed for carbapenem resistant strains to evaluate carbapenemase. Among 138 (91.4%) imipenem resistant A. baumannii isolates, 71 (47%) showed carbapenemase production by MHT (Figure 2). bla OXA-51 was surveyed and determined in total strains. Prevalence of bla TEM and bla PER genes among class A ESBL-producing A. baumannii isolates was 55 (36.4%) and 38 (25.2%), respectively (Figure 3). Fortunately, bla NDM-1 gene was not detected in the isolates (Figure 3).
ESBL Detection in Acinetobacter baumannii by Combination Disk Test (CDT) Method
MHT with Ertapenem Disks; 1, Quality control (MHT Positive K. pneumoniae ATCC BAA -1705); 2, Positive result, A positive MHT indicates that this isolate is producing a carbapenemase; 3, MHT negative result for carbapenemase detection by MHT.
PCR Amplified Products in This Study; Lane 1, DNA ladder 100 bp; lane 2, OXA -51 (353bp); lane 4, bla PER (925 bp); lane 6, control of bla NDM-1 (621 bp); lane 9, bla TEM (800 bp); lane 11, DNA ladder 100 pb.
5. Discussion
Acinetobacter baumannii is an important and common opportunistic nosocomial pathogen with high morbidity and mortality (13). The major cause for resistance to beta lactam antibiotics in A. baumannii is beta lactamase enzymes with genes such as TEM, PER, and OXA. In addition, resistance rate to some first line antibiotics including carbapenems is raising in the world as well as in our country (4, 14). Carbapenems activity of spectrum against most bacteria is broad, because these antibiotics relatively resist to hydrolysis via many of beta lactamases (4, 5). However, development of resistance mechanisms to most antimicrobial agents in this organism is a reason for raising multiple drug resistant pathogens in recent years (13).
In this study, 82.5% of the strains were A. baumannii and the rest were of other species of Acinetobacter. The most frequently isolated bacteria were obtained from tracheal aspirate followed by blood and the lowest frequency belonged to the bacteria isolated from pleural fluid and abscess. According to our research, the highest resistance rates of isolates were to ciprofloxacin, ceftazidime, amikacin, meropenem, and piperacillin and the lowest resistance rate denoted to colistin. Based on other investigations, it is obvious that appearance of resistant A. baumannii strains is increasing in the world (14). Our findings are consistent with the results of other earlier studies. Goudarzi et al. in 2013 reported that resistance rates of A. baumannii isolates were 99% to ceftazidime, meropenem, cefepime, 98% to ciprofloxacin, 91.5% to imipenem, and 70% to amikacin in Tehran (15). In addition, a study by Baygloo et al. in 2015 showed that all the A. baumannii isolates were resistant to cephalosporins and imipenem, but amikacin and piperacillin were sufficient to treat some of the patients (16).
In this study, two isolates (1.3%) were resistant to colistin. In a study conducted in East china by Zhao et al. in 2015, a high resistant rate was shown against ciprofloxacin (98.5%), ceftazidime (92.3%), imipenem (92.3%), and gentamycin (87.7 %) (17). It is possible that different times of these surveys are the cause of different outcomes of the studies. In our survey, CDT results indicated that 2 isolates (1.33%) of A. baumannii were positive for production of ESBLs. Results of CDT test were confirmed by PCR for different beta-lactamases genes including TEM and PER. However, 75 (49.7%) isolates included at least one of the Ambler class A beta lactamase genes by PCR. In the molecular method, 36.42% and 25.16% of the isolates were positive for bla TEM and bla PER, respectively. 18 isolates were positive for both β-lactamase genes (Figure 3). Several studies revealed the susceptibility of some strains of A. baumannii to beta-lactamase inhibitor clavulanic acid. Other investigations indicated that the results of this susceptibility test could yield false ESBL detection in A. baumannii (18).
Carbapenem resistant clinical isolates were screened by MHT test, showing that 71 isolates (47%) of A. baumannii were carbapenemase positive. A relatively similar study conducted by Kumar et al. (19) showed that 71% of isolates were carbapenemase positive by MHT. The results of this study are in contrast to a study reporting the rate of 14.8% by John et al. from India (20). The bla OXA-51 is a natural chromosomal gene of A. baumannii isolates. Carbapenem resistance may be related to bla OXA-51 gene in the strains of this bacteria (21) in this investigation in which, all clinical isolates carried bla OXA -51 gene and were determined as A. baumannii. In another study, bla OXA -51 gene was determined in all A. baumannii isolates from infectious patients (16, 22).
In the current research, the prevalence of bla TEM and bla PER genes in ESBL A. baumannii strains was 55 (36.42%) and 38 (25.16%), respectively. In addition, bla NDM-1 gene was not detected in A. baumannii strains. According to this study, TEM is the most prevalent ESBL genotype amongst A. baumannii strains that is inconsistent with other investigations that demonstrated various prevalence rates for TEM and PER. Zhao et al. showed that 25% of multidrug resistant isolates were positive for bla TEM in East china (13). A study conducted by Ahangarzadeh Rezaee et al. showed that the prevalence of Ambler classes A β-lactamase genes including TEM and PER was 37% (23). Bagheri et al. reported bla PER-1 in 52.4% of isolates in their research (24). Another study conducted in Iran showed that none of the A. baumannii isolates carried NDM gene (23, 25).
6. Conclusions
The universe appearance of broad-spectrum antibiotic resistance particularly to carbapenems in A. baumannii strains is a serious concern. The growth of antimicrobial resistance in bacteria can be prevented by wise prescribing. However, molecular genotyping studies for epidemiological surveillance of resistance genes in A. baumannii clinical strains can prevent the spread of these resistant bacteria and improve control strategies in particular areas.
Acknowledgements
References
-
1.
Routray A, Lavanya P, Soniya R, Madhavan R. Multiplex PCR for genes encoding prevalent OXA and NDM-1 carbapenemases in Acinetobacter. J Pharmacy Res. 2013;7(4):324-6. https://doi.org/10.1016/j.jopr.2013.03.026.
-
2.
Naas T, Bogaerts P, Bauraing C, Degheldre Y, Glupczynski Y, Nordmann P. Emergence of PER and VEB extended-spectrum beta-lactamases in Acinetobacter baumannii in Belgium. J Antimicrob Chemother. 2006;58(1):178-82. [PubMed ID: 16670107]. https://doi.org/10.1093/jac/dkl178.
-
3.
Sechi LA, Karadenizli A, Deriu A, Zanetti S, Kolayli F, Balikci E, et al. PER-1 type beta-lactamase production in Acinetobacter baumannii is related to cell adhesion. Med Sci Monit. 2004;10(6):BR180-4. [PubMed ID: 15173664].
-
4.
Bush K, Jacoby GA. Updated functional classification of beta-lactamases. Antimicrob Agents Chemother. 2010;54(3):969-76. [PubMed ID: 19995920]. https://doi.org/10.1128/AAC.01009-09.
-
5.
Opazo A, Dominguez M, Bello H, Amyes SG, Gonzalez-Rocha G. OXA-type carbapenemases in Acinetobacter baumannii in South America. J Infect Dev Ctries. 2012;6(4):311-6. [PubMed ID: 22505439].
-
6.
European Centre for Disease Prevention and Control. Carbapenem-resistant Acinetobacter baumannii in healthcare settings - 8 December 2016. Stockholm: ECDC; 2016.
-
7.
Shamsizadeh Z, Nikaeen M, Nasr Esfahani B, Mirhoseini SH, Hatamzadeh M, Hassanzadeh A. Detection of antibiotic resistant Acinetobacter baumannii in various hospital environments: potential sources for transmission of Acinetobacter infections. Environ Health Prev Med. 2017;22:44. https://doi.org/10.1186/s12199-017-0653-4.
-
8.
Mahon CR, Lehman DC, Manuselis G. Textbook of Diagnostic microbiology M. 5th ed. Pennsylvania: Elsevier Mosby; 2015. p. 484-5.
-
9.
Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol. 2006;44(8):2974-6. [PubMed ID: 16891520]. https://doi.org/10.1128/JCM.01021-06.
-
10.
CLSI. Performance standard for antimicrobial susceptibility testing: Twenty-fourth informational supplement. ClSL document M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute; 2014.
-
11.
Shahcheraghi F, Nobari S, Rahmati Ghezelgeh F, Nasiri S, Owlia P, Nikbin VS, et al. First report of New Delhi metallo-beta-lactamase-1-producing Klebsiella pneumoniae in Iran. Microb Drug Resist. 2013;19(1):30-6. [PubMed ID: 22984942]. https://doi.org/10.1089/mdr.2012.0078.
-
12.
Nordmann P, Poirel L, Carrer A, Toleman MA, Walsh TR. How To Detect NDM-1 Producers. J Clinical Microbiol. 2010;49(2):718-21. https://doi.org/10.1128/jcm.01773-10.
-
13.
Naiemi NA, Duim B, Savelkoul PH, Spanjaard L, de Jonge E, Bart A, et al. Widespread transfer of resistance genes between bacterial species in an intensive care unit: implications for hospital epidemiology. J Clin Microbiol. 2005;43(9):4862-4. [PubMed ID: 16145160]. https://doi.org/10.1128/JCM.43.9.4862-4864.2005.
-
14.
Feizabadi MM, Fathollahzadeh B, Taherikalani M, Rasoolinejad M, Sadeghifard N, Aligholi M, et al. Antimicrobial susceptibility patterns and distribution of blaOXA genes among Acinetobacter spp. Isolated from patients at Tehran hospitals. Jpn J Infect Dis. 2008;61(4):274-8. [PubMed ID: 18653968].
-
15.
Goudarzi H, Douraghi M, Ghalavand Z, Goudarzi M. Assessment of antibiotic resistance pattern in Acinetobacter bumannii carrying bla OXA type genes isolated from hospitalized patients. Novel Biomedicine. 2013;1(2):54-61.
-
16.
Shaykh Baygloo N, Bouzari M, Rahimi F, Abedini F, Yadegari S, Soroushnia M, et al. Identification of Genomic Species of Acinetobacter Isolated from Burns of ICU Patients. Arch Iran Med. 2015;18(10):638-42. [PubMed ID: 26443247].
-
17.
Zhao SY, Jiang DY, Xu PC, Zhang YK, Shi HF, Cao HL, et al. An investigation of drug-resistant Acinetobacter baumannii infections in a comprehensive hospital of East China. Ann Clin Microbiol Antimicrob. 2015;14:7. [PubMed ID: 25643932]. https://doi.org/10.1186/s12941-015-0066-4.
-
18.
Beceiro A, Fernandez-Cuenca F, Ribera A, Martinez-Martinez L, Pascual A, Vila J, et al. False extended-spectrum beta-lactamase detection in Acinetobacter spp. due to intrinsic susceptibility to clavulanic acid. J Antimicrob Chemother. 2008;61(2):301-8. [PubMed ID: 18065824]. https://doi.org/10.1093/jac/dkm461.
-
19.
Kumar AV, Pillai VS, Dinesh KR, Karim S. The phenotypic detection of Carbapenemase in meropenem resistant Acinetobacter calcoaceticus baumannii complex in a tertiary care hospital in south India. J Clin Diagn Res. 2011;5:223-6.
-
20.
John S, Balagurunathan R. Metallo beta lactamase producing Pseudomonas aeruginosa and Acinetobacter baumannii. Indian J Med Microbiol. 2011;29(3):302-4. [PubMed ID: 21860115]. https://doi.org/10.4103/0255-0857.83918.
-
21.
Heritier C, Poirel L, Nordmann P. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin Microbiol Infect. 2006;12(2):123-30. [PubMed ID: 16441449]. https://doi.org/10.1111/j.1469-0691.2005.01320.x.
-
22.
Azimi L, Talebi M, Pourshafie MR, Owlia P, Rastegar Lari A. Characterization of Carbapenemases in Extensively Drug Resistance Acinetobacter baumannii in a Burn Care Center in Iran. Int J Mol Cell Med. 2015;4(1):46-53. [PubMed ID: 25815282].
-
23.
Rezaee MA, Pajand O, Nahaei MR, Mahdian R, Aghazadeh M, Ghojazadeh M, et al. Prevalence of Ambler class A beta-lactamases and ampC expression in cephalosporin-resistant isolates of Acinetobacter baumannii. Diagn Microbiol Infect Dis. 2013;76(3):330-4. [PubMed ID: 23726148]. https://doi.org/10.1016/j.diagmicrobio.2013.04.003.
-
24.
Bagheri Josheghani S, Moniri R, Firouzeh F, Sehat M, Dasteh Goli Y. Susceptibility pattern and distribution of oxacillinases andblaPER-1 genes among multidrug resistant Acinetobacter baumannii in a teaching hospital in Iran. J Pathog. 2015;2015:7. https://doi.org/10.1155/2015/957259.
-
25.
Alan TF, Goudarzi H, Fallah F, Hashemi A, Doustdar F, Bostan H. Detection of blaNDM, blaDIM, blaIMP, blaVIM and blaCTX-M-15 beta-lactamase genes among pseudomonas aeruginosa and acinetobacter baumannii strains isolated from two hospitals of Tehran, Iran. Novel Biomed. 2016;4(4):153-8.
