Abstract
Background:
Malaria, as a parasitic disease, is one of the most important public health problems in Iran. Malaria is mainly diagnosed by peripheral blood smear, stained by Giemsa; in Iran it is also diagnosed by blood smear that is highly depends on technician’s skills and laboratory properties.Objectives:
Correct diagnosis of malaria and identification of human malaria species in spite of the measures taken to eliminate the disease in Iran, have made the complete understanding of malaria epidemiology critical. Therefore, the current study aimed at investigating the epidemiology of 2 species of human Plasmodium in Sistan and Baluchistan province using polymerase chain reaction (PCR) method.Methods:
The present descriptive study was conducted on 100 patients suspected to malaria infection who referred to health centers of Chabahar, Iranshahr, Nikshahr, and Sarbaz districts. DNAs were extracted from blood samples using the specific kit, and nested-PCR reaction was performed to identify the Plasmodium species according to NP-2013 protocol.Results:
Molecular analysis was performed on 100 samples suspected of malaria; 84 negative and 16 positive samples were detected including 8 Plasmodium vivax, 2 P. falciparum, and 6 mixed infections (P. vivax and P. falciparum). No P. ovale or P. malariae was observed.Conclusions:
The results showed that malaria had a decreasing trend in Sistan and Baluchistan province. Therefore, the malaria elimination program is applicable and attainable in this region as a goal.Keywords
Malaria Plasmodium Species Nested-PCR Iran Plasmodium vivax P. falciparum
1. Background
In the last decade, malaria control progressed globally (1, 2). Recently, the world health organization (WHO) sets a mission on malaria elimination in West Asia and Southeast Asia in order to control this threat (2). Malaria is caused by eukaryotic microorganism from genus Plasmodium with 6 human-specific species including Plasmodium falciparum, P. vivax, P. malariae, P. knowlesi, P. ovale wallikeri, and P. ovale curtisi (3, 4). Most of malaria cases in Iran are reported from Sistan and Baluchistan, Kerman, and Hormozgan provinces (5).
The simple and efficient microscopic diagnostic methods are the golden standards to diagnose malaria and are routinely used in laboratories. However, there are some errors in the identification of Plasmodium species, which sometimes lead to false negative especially in low rate parasitemia (6). One consistent technique to evaluate the malaria immune response is to study the human antibodies produced during the infection. Evaluation of these antibodies in serological studies by recombinant antigens is commonly recognized valuable to increase parasitological documents as well as transmission and immunity of malaria in the areas evaluated (7).
One of the examinations commonly used in epidemiological and diagnostic tests is the enzyme-linked immunosorbent assay (ELISA), sensitivity and specificity of which is considerable. This technique can identify IgM and IgG specific antibodies and also test a large number of samples at the same time (8). Thus, recognizing and evaluating the potentiality of immunodominant P. vivax diagnostic antigens to be used in ELISA test are significant and beneficial. But in the blood stages, specific antibodies may be identified and improved to high levels following clinical signs that are advantages of parasite presence in the patient’s blood, which continue nearly for 6 months or seldom longer, and then, decline to inactive levels (9, 10). Moreover, it is not desirably possible to identify concurrent presence of 2 or more strains of Plasmodium in the sample or identify parasite after drug administration through microscopic methods and also in low rate parasitemia or reduction of parasite rate due to drug consumption (11).
According to the critical role of early identification and diagnosis of the parasite and also its subspecies, especially in asymptomatic and low parasitemia in prevention, control and eradication of malaria programs, molecular identification methods are necessary and unavoidable for better isolation and higher identification precision, particularly for the isolation of new human Plasmodium spp. such as P. ovale wallikeri and P. ovale curtisi. Thus, nested-polymerase chain reaction (PCR) was used to identify the isolated Plasmodium species as a cause of malaria. In nested-PCR technique, 2 consequent PCRs were performed and the product of the 1st PCR was used as the template for the 2nd PCR. Whenever the isolated DNA was low, this procedure was used to improve the sensitivity of the test. The nested-PCR has higher sensitivity to identify Plasmodium spp. in asymptomatic cases and also to differentiate Plasmodium species than regular PCR; and could identify 1 to 10 parasites in each microliter of a blood sample (12).
2. Objectives
The current study aimed at testing the sensitivity and specificity of nested-PCR to detect malaria in South-East of Iran with assortment of detailed epidemiological statistics to study the prevalence and risk factors for malaria contact.
3. Methods
3.1. Ethics Statement
The current qualitative study was conducted with the purpose of epidemiologic investigation of different types of malaria in Sistan and Baluchistan province. The experimental design was performed in accordance with the current ethical norms approved by Zahedan University of Medical Sciences ethical committee acts (IR.Zamus.IR.1395.2; proposal code 1818), based on the specific national ethical guidelines for biomedical research issued by the research and technology deputy of ministry of health and medical education (MOHME) of Iran.
3.2. Sampling
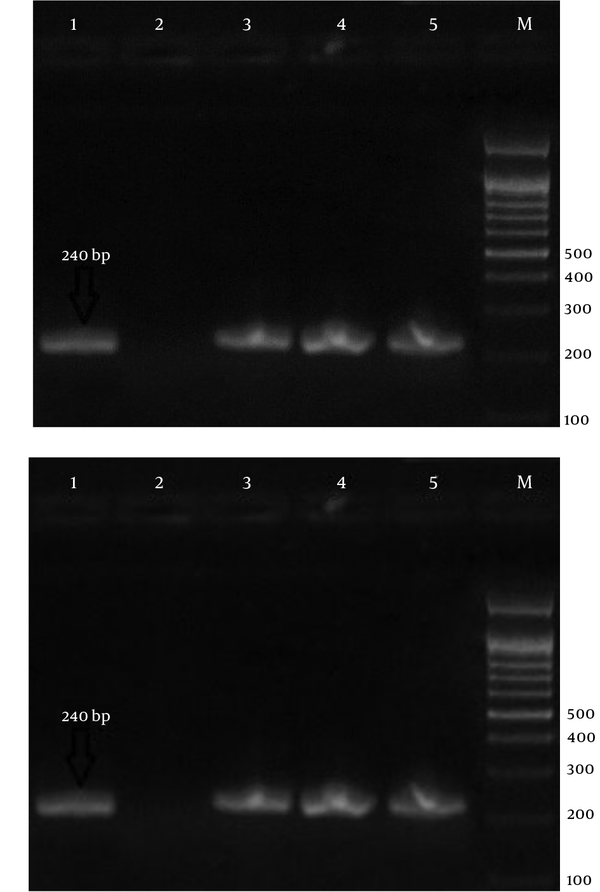
A total of 100 malaria-suspected samples including Iranian and foreigners were collected from 4 districts of Nikshahr, Iranshahr, Sarbaz, and Chahbahar from January 2015 to February 2016 and transferred to the research center of Bualisina hospital. The DNA extraction was conducted using DNA-extraction kit from blood and flesh – bar method (DynaBio Blood/Tissue Genomic DNA Extraction Kit). The nested-PCR technique was carried out to identify Plasmodium species using the designed primers (Table 1). A pair of oligonucleotide primers (rPLU1 and rPLU5, CinnaGen Company; Iran) were used for PCR-1 reaction. Then, in PCR-2, the first primers were applied (rPLU3 and rPLU4), and then, to confirm the presence of Plasmodium spp. in the suspected samples, samples that were positive for the above mentioned primers and showed corresponding bands (240 bp) were treated using identification primers (1. rFAL, 2. rFAL, 1. rVIV, 2. rVIV, 1. rMAL, 2. rMAL, 1. rOVAL, 2. rOVAI; CinnaGen, Iran). The primary PCR product was diluted using distilled water with proportion of 1:100 and used as template in the secondary PCR.
Primers Used in the Current Study
| Primer | Sequence (5 = -3 =) | Species/Genus | bp | Reference |
|---|---|---|---|---|
| rPLU1 | TCA AAG ATT AAG CCA TGC AAG TGA | Genus-nest 1 | _1,670 | (13) |
| rPLU5 | CCT GTT GTT GCC TTA AAC TTC | |||
| rPLU3 | TTT TTA TAA GGA TAA CTA CGG AAA AGCTGT | Genus-nest 2 | 240 | (13) |
| rPLU4 | TAC CCG TCA TAG CCA TGT TAG GCC AAT ACC | |||
| rFAL1 | TTA AAC TGG TTT GGG AAA ACC AAA TAT ATT | P. falciparum | 206 | (13) |
| rFAL2 | ACA CAA TGA ACT CAA TCA TGA CTA CCC GTC | |||
| rMAL1 | ATA ACA TAG TTG TAC GTT AAG AAT AAC CGC | P. malaria | 145 | (13) |
| rMAL2 | AAA ATT CCC ATG CAT AAA AAA TTA TAC AAA | |||
| rVIV1 | CGC TTC TAG CTT AAT CCA CAT AAC TGA TAC | P. vivax | 121 | (13) |
| rVIV2 | ACT TCC AAG CCG AAG CAA AGA AAG TCC TTA | |||
| rOVA1WC | TGT AGT ATT CAA ACG CAG T | P. ovale sp. | 659 - 662 | (14) |
| rOVA2WC | TAT GTA CTT GTT AAG CCT TT | |||
| rOVA1 | ATC TCT TTT GCT ATT TTT TAG TAT TGG AGA | P. ovale curtisia | 787 - 789 | (15) |
| rOVA2 | ATC TAA GAA TTT CAC CTC TGA CAT CTG | |||
| rOVA1v | ATC TCC TTT ACT TTT TGT ACT GGA GA | P. ovale wallikeria | 782 | (16) |
| rOVA2v | GGA AAA GGA CAC TAT AAT GTA TCC TAA TA |
The amplification conditions were as follows: initial denaturing at 95°C for 5 minutes; 25 cycles at 94°C for 60 seconds, 58°C for 60 seconds, and 72°C for 2 minutes; final elongation was conducted at 72°C for 7 minutes. End of the reaction and reduction of temperature was conducted at 20°C. The thermocycler temperature and time conditions were the same for the 1st step of the nested-PCR and the 2nd step of species identification nested-PCR. However, in the 2nd step all the tests were similar except for the cycles 2 to 4; totally, 25 cycles were run in the 1st step and 30 cycles in the 2nd step to identify P. vivax, P. falciparum, and P. malaria, (13, 14).
In order to identify P. ovale, the following steps were carried out: initial denaturing at 95°C for 5 minutes; 30 cycles at 94°C for 60 seconds, 64°C for 60 seconds, and 72°C for 2 minutes; final elongation was run at 72°C for 5 minutes. End of the reaction and reduction of temperature were at 20°C. The nested-PCR products were analyzed by electrophoresis on 1.5% agarose gel, stained with red gel (15, 16) (Figures 1 - 4).
Electrophoresis of PCR Products Using Strain-Specific Oligonucleotide ssrRNA to Detect Malaria Parasites in the Samples; a 100-bp Marker Was Used.
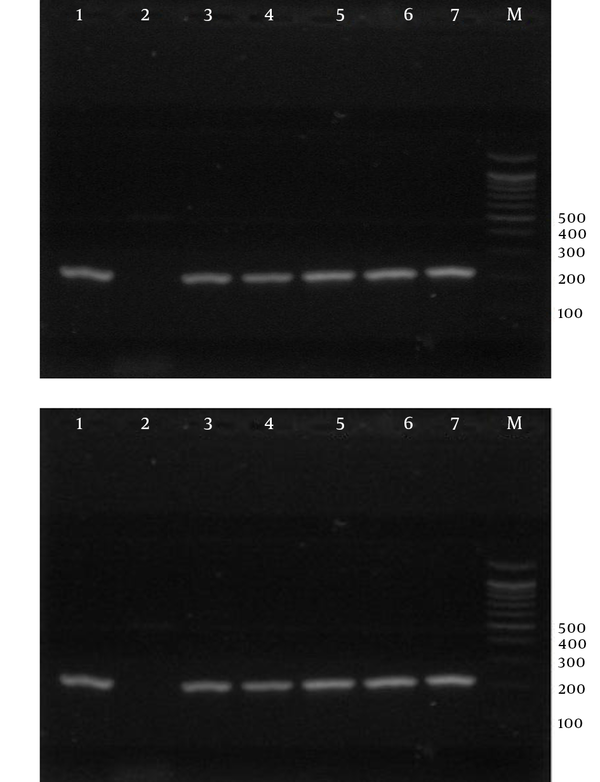
Electrophoresis of PCR Products Using Strain-Specific Oligonucleotide ssrRNA to Detect Plasmodium falciparum in the Samples; a 100-bp Marker Was Used.
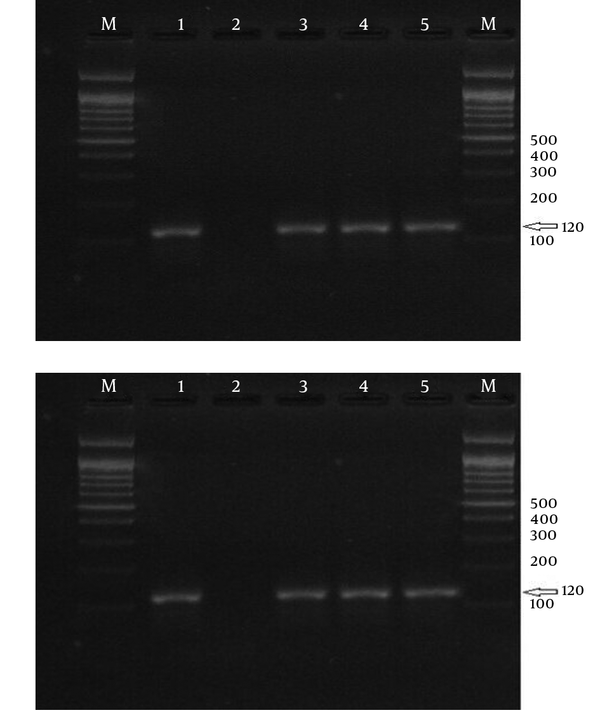
Electrophoresis of PCR Products Using Strain-Specific Oligonucleotide ssrRNA to Detect Plasmodium vivax in the samples; a 100-bp Marker Was Used.
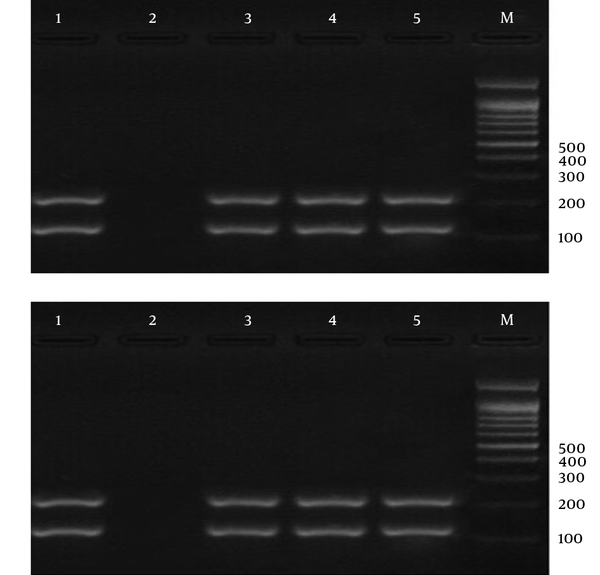
Electrophoresis of PCR Products Using Strain-Specific Oligonucleotide ssrRNA to Detect Mixed Infection (Plasmodium vivax and Plasmodium falciparum) in the Samples; a 100-bp Marker Was Used.
4. Results
Based on the 100 malaria-suspected samples, 43 males (43%) and 57 females (57%), 86 Iranian (86%) and 14 foreigners (14%) (Afghan and Pakistani mainly) aged 8 to 60 years, 84 samples were negative and 16 were positive out of which 2 P. falciparum and 14 P. vivax were detected by the microscopic examination. But, molecular analysis detected 8 P. vivax, 2 P. falciparum, and 6 mixed samples (P. vivax and P. falciparum). Six mixed cases were diagnosed as P. vivax in the primary laboratory diagnosis, although no P. ovale and P. malariae were observed. Interestingly, the disease was observed only in Iranian cases.
5. Discussion
Laboratory diagnosis of malaria is currently performed to detect parasites by the light microscopy of Geimsa-stained thick and thin blood smears. This procedure is cheap and simple, but is a labor-intensive procedure, which requires well-trained personnel (17). When parasitemia is very low, the data of microscopy diagnosis are limited, and in some cases biased by the inability to devote the necessary amount of time to the examination of blood smears (17).
Greater sensitivity and specificity of PCR method in comparison with the thick blood films examination method are shown by many studies. The detection of low P. vivax and P. falciparum parasitemia by PCR, at levels undetectable by microscopy, was reported earlier (14). The current study on microscopy and nested-PCR assay showed that the results obtained by PCR were equivalent or superior to those of the microscopy, in which all microscopy-positive samples were positive by PCR. In addition, the PCR test was able to detect mix infections missed by microscopy. This may be due to the tendency of 1 species to be dominant over other ones (14, 18).
Given the progresses achieved over 5 decades of working on malaria eradication and also significant reduction in the incidence of the disease in the recent 10 years, complete elimination of malaria in the country is the main agenda of the joint direction of the Iranian ministry of health and medical education and WHO. Nowadays, the local distribution and transmission of malaria in Iran is limited to 28 cities, located in Sistan and Baluchistan, Hormozgan, and Kerman provinces. The ultimate goal of malaria elimination program in 2025 horizon is interrupting local transmission of the disease in Iran (19-23).
Malaria elimination is defined as halting its transmission by anopheles mosquitoes in a specific geographical area to reach 0 incidence of the disease. Malaria eradication is defined as continuous and persistent reduction in the global incidence of malaria until reaching the rate of zero (24). According to WHO reports, malaria is on the verge of elimination in Iran (25). Iran reported only 330 cases of malaria in 2015, which is about 50% reduction compared with 2014 (26). Malaria is a major health problem in the Southeastern regions of Iran and greatly inhibits the socioeconomic development of the region. In the current study, nested-PCR molecular technique was used, which has high sensitivity; it can detect mixed infectious agents as well as parasite species. Many studies showed the high sensitivity and specificity of PCR, compared with those of the microscopic methods (27-31).
Zakeri et al. investigated the malaria infection prevalence in the Southeastern areas of Iran using nested-PCR method. They tested 120 samples targeting ssrRNA gene using molecular methods. They identified P. vivax in 59%, P. falciparum in 10%, and a mix infection of P. vivax and P. falciparum in 28.4% of the samples (15). Ebrahimzadeh et al. studied malaria strains by molecular methods. They analyzed 140 samples, 94.3% of which were positive; 51.4% were P. vivax; 12.6% P. falciparum and 29.3% were a mixed infection of P. vivax and P. falciparum (32). Plasmodium malaria was also reported (33).
Zakeri et al. investigated the sensitivity of nested-PCR in the diagnosis of a mixed infection of P. falciparum and P. vivax in Pakistan, Iran, and Afghanistan. They studied 100 P. vivax-positive samples diagnosed by microscopic methods in Iran and mix-infections in 22% of the samples (34). Different studies in Iran reported a reduction in malaria incidence in recent years (35-39). The current research investigated malaria species via molecular methods; the results revealed that out of 100 malaria-suspected samples, 84% were negative and 16% positive. The positive samples included 8% P. vivax, 2% P. falciparum, and 6% of positive samples were a mix-infection of P. falciparum and P. vivax. However, P. vivax was the dominant species in the current study, which was consistent with previous studies. No P. ovale or P. malaria species was observed; as found by Ebrahimzadeh et al. (40).
Microscopic methods are the golden standards for the simple and efficient identification of malaria, which conduct routinely in medical laboratories; but there is potential for error to identify parasite type and false negative reports in low-rate parasitemia (asymptomatic malaria). Moreover, in microscopic methods, there is no possibility to identify mixed plasmodium infection or diagnose the disease after drug consumption due to low parasite count. Furthermore, the presence of mixed-infection is an important factor for disease severity; the presence of P. vivax can have suppressing effects on P. falciparum infection, if it remains undiagnosed. On the other hand, considering the necessity of special treatment for P. falciparum infection, mixed infection including this species can lead to increase in P. falciparum infected cases that increases the number of parasite carriers in the population, and therefore, increases the incidence of the disease.
5.1.Conclusion
Correct diagnosis of malaria species can help proper treatment of the disease and reduce carriers in the general population, and accordingly, reduce the risk of malaria infection. According to the goal of malaria control program to minimize mortality rate from malaria and decrease the prevalence of the disease, a reliable laboratory test is necessary for the early diagnosis of the disease. The current study findings suggested that PCR diagnostic method was very helpful in the areas where the mixed infection of P. falciparum and P. vivax is common. It can contribute to precise determination of incidence rate of the disease and is also useful in follow-up endeavors, as a very useful supplement for microscopic diagnosis of malaria. Unfortunately, there is no facility or procurement to perform PCR tests in rural areas; therefore, every 5 years epidemiological molecular studies can narrow this gap. Moreover, considering the previous studies and the current study findings, it is apparent that malaria had a declining trend in recent years in Baluchistan. Thus, implementation of malaria elimination program is feasible and attainable in the province.
Acknowledgements
References
-
1.
Murray CJ, Rosenfeld LC, Lim SS, Andrews KG, Foreman KJ, Haring D, et al. Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet. 2012;379(9814):413-31. [PubMed ID: 22305225]. https://doi.org/10.1016/S0140-6736(12)60034-8.
-
2.
World malaria report 2014. Geneva: World Health Organization; 2016. Available from: http://www.who.int/malaria/publications/world_malaria_report_2014/wmr-2014-no-profiles.pdf.
-
3.
Chew CH, Lim YA, Lee PC, Mahmud R, Chua KH. Hexaplex PCR detection system for identification of five human Plasmodium species with an internal control. J Clin Microbiol. 2012;50(12):4012-9. [PubMed ID: 23035191]. https://doi.org/10.1128/JCM.06454-11.
-
4.
Fuehrer HP, Noedl H. Recent advances in detection of Plasmodium ovale: implications of separation into the two species Plasmodium ovale wallikeri and Plasmodium ovale curtisi. J Clin Microbiol. 2014;52(2):387-91. [PubMed ID: 24478466]. https://doi.org/10.1128/JCM.02760-13.
-
5.
World Malaria Risk Chart. International Association for Medical Assistance to Travellers Guelph. Canada; 2014. Available from: http://www.iamat.org.
-
6.
Alemu A, Fuehrer HP, Getnet G, Kassu A, Getie S, Noedl H. Comparison of Giemsa microscopy with nested PCR for the diagnosis of malaria in North Gondar, north-west Ethiopia. Malar J. 2014;13:174. [PubMed ID: 24884606]. https://doi.org/10.1186/1475-2875-13-174.
-
7.
Mirahmadi H, Spotin A, Fallahi S, Taghipour N, Turki H, Seyyed Tabaei SJ. Cloning and Sequence Analysis of Recombinant Plasmodium vivax Merozoite Surface Protein 1 (PvMSP-142 kDa) In pTZ57R/T Vector. Iran J Parasitol. 2015;10(2):197-205. [PubMed ID: 26246817].
-
8.
Mirahmadi H, Fallahi S, Omrani VF, Kazemi B, Haghighi A, Tabaei SJ. High-Level Expression of Immunogenic Recombinant Plasmodium vivax Merozoite Surface Protein (Pvmsp-142 kDa) in pGEX 6P1 Vector. Iran J Public Health. 2015;44(1):89. [PubMed ID: 26060780].
-
9.
Voller A, Meuwissen JHE, Verhave H. Methods of measuring the immunological response to plasmodia. In: Kreier JP, editor. Immunology and Immunisation. New York: Malaria Academic Press; 1980. p. 67-109.
-
10.
Mirahmadi H, Fallahi S, Seyyed Tabaei SJ. Soluble recombinant merozoite surface antigen-142kDa of Plasmodium vivax: An improved diagnostic antigen for vivax malaria. J Microbiol Methods. 2016;123:44-50. [PubMed ID: 26851675]. https://doi.org/10.1016/j.mimet.2016.02.003.
-
11.
Barber BE, William T, Grigg MJ, Yeo TW, Anstey NM. Limitations of microscopy to differentiate Plasmodium species in a region co-endemic for Plasmodium falciparum, Plasmodium vivax and Plasmodium knowlesi. Malar J. 2013;12:8. [PubMed ID: 23294844]. https://doi.org/10.1186/1475-2875-12-8.
-
12.
Singh B, Cox-Singh J, Miller AO, Abdullah MS, Snounou G, Rahman HA. Detection of malaria in Malaysia by nested polymerase chain reaction amplification of dried blood spots on filter papers. Trans R Soc Trop Med Hyg. 1996;90(5):519-21. [PubMed ID: 8944260].
-
13.
Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61(2):315-20. [PubMed ID: 8264734].
-
14.
Snounou G, Singh B. Nested PCR analysis of Plasmodium parasites. Methods Mol Med. 2002;72:189-203. [PubMed ID: 12125116]. https://doi.org/10.1385/1-59259-271-6:189.
-
15.
Zakeri S, Najafabadi S, Zare A, Djadid ND. Detection of malaria parasites by nested PCR in south-eastern, Iran: Evidence of highly mixed infections in Chahbahar district. Malaria J. 2002;1(1):2. https://doi.org/10.1186/1475-2875-1-2.
-
16.
Brown AE, Kain KC, Pipithkul J, Webster HK. Demonstration by the polymerase chain reaction of mixed Plasmodium falciparum and P. vivax infections undetected by conventional microscopy. Trans R Soc Trop Med Hyg. 1992;86(6):609-12. [PubMed ID: 1287912].
-
17.
Calderaro A, Piccolo G, Perandin F, Gorrini C, Peruzzi S, Zuelli C, et al. Genetic polymorphisms influence Plasmodium ovale PCR detection accuracy. J Clin Microbiol. 2007;45(5):1624-7. [PubMed ID: 17360843]. https://doi.org/10.1128/JCM.02316-06.
-
18.
Fuehrer HP, Stadler MT, Buczolich K, Bloeschl I, Noedl H. Two techniques for simultaneous identification of Plasmodium ovale curtisi and Plasmodium ovale wallikeri by use of the small-subunit rRNA gene. J Clin Microbiol. 2012;50(12):4100-2. [PubMed ID: 23015675]. https://doi.org/10.1128/JCM.02180-12.
-
19.
Malaria elimination: a field manual for low and moderate endemic countries. Geneva: World Health Organization; 2007. Available from: http://apps.who.int/iris/bitstream/10665/43796/1/9789241596084eng.pdf.
-
20.
Global technical strategy for malaria 2016-2030. Geneva: World Health Organization; 2015. Available from: https://www.google.com/?gws_rd=ssl#q=Global+technical+strategy+for+malaria+2016-2030.
-
21.
Guidelines on prevention of the re-introduction of malaria. World Health Organization Regional Office for the Eastern Mediterranean. Cairo; 2007. Available from: http://applications.emro.who.int/dsaf/dsa743.pdf.
-
22.
From malaria control to malaria elimination: A manual for elimination scenario planning. Global Malaria Programme. Geneva: World Health Organization; 2014. Available from: http://apps.who.int/iris/bitstream/10665/112485/1/9789241507028_eng.pdf.
-
23.
National Strategic Plan for Malaria Control, Islamic Republic of Iran (2004-2008). 2004.
-
24.
Lines J, Whitty CJM, Hanson K. Prospects for Eradication and Elimination of Malaria: a technical briefing for DFID. 2007.
-
25.
Elimination. 2016. Available from: http://www.who.int/malaria/areas/elimination/overview/en/.
-
26.
Field guide for malaria epidemic assessment and reporting. Geneva: World Health Organization; 2004. Available from: http://apps.who.int/iris/bitstream/10665/68764/1/WHOHTMMAL2004.1097.pdf.
-
27.
Singh N, Shukla MM, Shukla MK, Mehra RK, Sharma S, Bharti PK, et al. Field and laboratory comparative evaluation of rapid malaria diagnostic tests versus traditional and molecular techniques in India. Malar J. 2010;9:191. [PubMed ID: 20602766]. https://doi.org/10.1186/1475-2875-9-191.
-
28.
Tham JM, Lee SH, Tan TM, Ting RC, Kara UA. Detection and species determination of malaria parasites by PCR: comparison with microscopy and with ParaSight-F and ICT malaria Pf tests in a clinical environment. J Clin Microbiol. 1999;37(5):1269-73. [PubMed ID: 10203469].
-
29.
Rodulfo H, De Donato M, Mora R, Gonzalez L, Contreras CE. Comparison of the diagnosis of malaria by microscopy, immunochromatography and PCR in endemic areas of Venezuela. Braz J Med Biol Res. 2007;40(4):535-43. [PubMed ID: 17401497].
-
30.
Coleman RE, Sattabongkot J, Promstaporm S, Maneechai N, Tippayachai B, Kengluecha A, et al. Comparison of PCR and microscopy for the detection of asymptomatic malaria in a Plasmodium falciparum/vivax endemic area in Thailand. Malar J. 2006;5:121. [PubMed ID: 17169142]. https://doi.org/10.1186/1475-2875-5-121.
-
31.
Nankabirwa JI, Yeka A, Arinaitwe E, Kigozi R, Drakeley C, Kamya MR, et al. Estimating malaria parasite prevalence from community surveys in Uganda: a comparison of microscopy, rapid diagnostic tests and polymerase chain reaction. Malar J. 2015;14:528. [PubMed ID: 26714465]. https://doi.org/10.1186/s12936-015-1056-x.
-
32.
Ebrahimzadeh A, Fouladi B, Fazaeli A. High rate of detection of mixed infections of Plasmodium vivax and Plasmodium falciparum in South-East of Iran, using nested PCR. Parasitol Int. 2007;56(1):61-4. [PubMed ID: 17257891]. https://doi.org/10.1016/j.parint.2006.12.001.
-
33.
Adel E, Asghar F. The risk of re-emergence of Plasmodium malariae in south-east of Iran as detected by nested polymerase chain reaction. Asian J Epidemiol. 2008;1(2):47-52.
-
34.
Zakeri S, Kakar Q, Ghasemi F, Raeisi A, Butt W, Safi N, et al. Detection of mixed Plasmodium falciparum & P. vivax infections by nested-PCR in Pakistan, Iran & Afghanistan. Indian J Med Res. 2010;132:31-5. [PubMed ID: 20693586].
-
35.
Amiri SA, Delavari A, Amiri A, Alipour H. Epidemiology of malaria in Nikshahr, Sistan and Baluchestan province, Southeast Iran, during 2004-2010. Ann Trop Med Public Health. 2013;6(4):430. https://doi.org/10.4103/1755-6783.127790.
-
36.
Ostovar A, Raeisi A, Haghdoost AA, Ranjbar M, Rahimi A, Sheikhzadeh K, et al. Lessons learnt from malaria epidemics in the Islamic Republic of Iran. East Mediterr Health J. 2012;18(8):864-9. [PubMed ID: 23057376].
-
37.
Sargolzaie, N, Salehi M, Kiani M, Sakeni M, Hasanzehi A. Malaria Epidemiology in Sistan and Balouchestan Province during April 2008-March 2011, Iran. ZJRMS. 2014;16(4):41-3.
-
38.
Reza YM, Taghi RM. Prevalence of malaria infection in Sarbaz, Sistan and Bluchistan province. Asian Pac J Trop Biomed. 2011;1(6):491-2. [PubMed ID: 23569820]. https://doi.org/10.1016/S2221-1691(11)60107-X.
-
39.
Raeisi A, Gouya MM, Nadim A, Ranjbar M, Hasanzehi A, Fallahnezhad M, et al. Determination of Malaria Epidemiological Status in Iran's Malarious Areas as Baseline Information for Implementation of Malaria Elimination Program in Iran. Iran J Public Health. 2013;42(3):326-33. [PubMed ID: 23641411].
-
40.
Ebrahimzadeh A, Mohammadi S, Polshekan M, Jamshidi A, Mehravaran A. Evaluation of negative malaria giemsa-stained smears referred from malaria centers in Sistan and Balouchestan using nested polymerase chain reaction. Zahedan J Res Med Sci. 2014;16(4):15-8.