Abstract
Background:
Urinary tract infection (UTIs) caused by Escherichia coli is one of the most common human complications. The discriminate use of antibiotics leads to increasing multiple-drug resistance (MDR) in E. coli.Objectives:
This study aimed at investigating the frequency of fluoroquinolones resistance by detecting qnr genes and mutation in gyrA gene in clinical isolates of E. coli in the North of Iran.Methods:
This cross-sectional study was conducted on 309 E. coli isolates collected from major hospitals in Rasht, North of Iran. All isolates were identified by standard microbiological methods. Antimicrobial susceptibility testing was conducted by the disk diffusion method for the three quinolone antibiotics. The detection of gyrA, qnrS, qnrB, and qnrA genes was performed by the polymerase chain reaction (PCR) method. PCR products of gyrA gene were digested by HinfI enzyme to identify the mutation in gyrA gene by restriction fragment length polymorphism (RFLP) method.Results:
Of 309 tested E. coli isolates, the most common resistance was observed against nalidixic acid (60.2%) and the least common one against norfloxacin (40.5%). The prevalence of qnrS, qnrB, and qnrA genes was 4.5%, 73.1%, and 62.8%, respectively. Mutation in gyrA gene was observed in codon 83 in 56.3% of the strains.Conclusions:
In summary, despite the significant association of the investigated genes with fluoroquinolones resistance in these bacteria, the presence of these genes in susceptible strains suggested that some other possible mechanisms may also influence resistance against fluoroquinolones.Keywords
1. Background
Urinary tract infections (UTIs) are one of the most common human complications, especially among females (1, 2). Several bacteria such as Enterobacteriaceae can cause this complication and Escherichia coli is among the most significant ones (3). This bacterium is responsible for nearly 90% of UTIs among females (4). Often, no single and specific treatment can be found. Sulfonamide, ampicillin, cephalosporins, fluoroquinolones, and aminoglycosides are helpful to treat UTIs (1, 4).
Although the antibiotic resistance rate of E. coli can differ in the diverse geographic area, the discriminate use of antibiotics leads to increasing multi-drug resistance in this microorganism (5, 6). In the last decade, antibiotics such as ampicillin and co-trimoxazole were used to treat infections caused by E. coli, but nowadays, quinolones such as nalidixic acid and ciprofloxacin are replaced. Quinolones are synthetic products with similar structure and mechanism of action (7). Nalidixic acid and some other older quinolones are replaced by the newer fluoroquinolones with broad-spectrum antibacterial activities. Quinolones selectively and reversibly inhibit DNA gyrase subunit A activity and induce the formation of relaxation complex analogue (1, 7). Resistance to quinolones is mainly mediated by a mutation in the subunit A of DNA gyrase on chromosomes (8). In Gram-negative bacteria, topoisomerase IV is a secondary target for quinolones. The qnr genes are responsible for plasmid-mediated resistance to quinolones and prevent the inhibitory effect of these antibiotics on DNA gyrase and topoisomerase (9, 10).
Determination of the resistance rate of E. coli strains to these antibiotics and prescription of proper curative antibiotics can lead to a decrease in the spread of antibiotic-resistant strains (11). Therefore, it is necessary to perform routine national or international surveillance to test antibiotic susceptibility, due to the increasing rate of antibiotic resistance, and extensive changes in the efficacy of some drugs.
2. Objectives
The present study aimed at determining the emergence of quinolone resistance and the prevalence of qnr genes, and mutation in gyrA gene in clinical isolates of E. coli isolated in the North of Iran.
3. Methods
3.1. Ethics Statement
This study was in accordance with the declaration of Helsinki and was approved by the local Ethics Committee (Approval No. IR.GUMS.REC.1394.343). All patients’ personal information was kept confidential.
3.2. Study Design and Samples
This cross-sectional study was performed from March 2014 to August 2014 on 1723 urine samples of patients with UTIs referred to major hospitals in the North of Iran. All urine samples were cultured on blood agar and Eosin methylene blue (EMB) agar plates and incubated for 24 - 48 hours at 37°C. Oxidase and catalase tests as well as Gram stain were performed on the grown colonies. The colonies were cultured in triple sugar iron agar (TSI), SIM (sulfur, indole, motility), MR-VP (methyl red-Voges Proskauer), urea, Simmon’s citrate, and lysine iron agar (LIA) media and incubated at 37°C for 24 hours. All culture media were purchased from Merck, Darmstadt, Germany.
3.3. Antimicrobial Susceptibility Testing
Antibiogram was performed on colonies of bacteria identified as E. coli by disk diffusion method using three antibiotic disks (MAST, UK) including ofloxacin, norfloxacin, and nalidixic acid according to Clinical & Laboratory Standards Institute (CLSI) guidelines. The diameter of the inhibition zone was measured, and the results were interpreted based on CLSI instructions (12).
3.4. Chromosomal and Plasmid DNA Extraction
In order to study mutation in gyrA gene and the frequency of qnr genes, all chromosomal and plasmids DNA of E. coli strains were extracted by two sets of kits including high pure DNA template preparation kit (Roche, Germany) and gene JET plasmid miniprep kit (Fermentas, Lithuania), respectively based on the manufacturers instructions.
3.5. Amplification of qnrS, qnrB, and qnrA Genes by Multiplex PCR
The primers were designed by PrimerPlex (PREMIER Biosoft). The designed primers were ordered to Bioneer Company in South Korea for synthesis. PCR reactions were performed in a final volume of 25 μL containing 10 pmol of each primer, 20 - 50 ng of extracted plasmid DNA, 200 μM of nucleoside triphosphate, and one unit of Taq DNA polymerase. Amplification was performed with pre-incubation (94ºC for four minutes), followed by 35 cycles of denaturation (94ºC for 15 seconds), annealing (60ºC for 10 seconds), extension (72ºC for 15 seconds) and a final extension (72ºC for 10 minutes). The list of used primers is shown in Table 1.
Primer Sequences Used for the Amplification of qnr and gyrA Genes
| Primer Designation | Primer Sequences | Size of Product, bp |
|---|---|---|
| qnrA | 319 | |
| qnrA-F | 5’ CCA GGA TTT GAG TGA CAG CCG TTT 3’ | |
| qnrA-R | 5’ ACC TGA GAT NATA AGC CGA GCA GAA G 3’ | |
| qnrB | 134 | |
| qnrB-F | 5’ GGC GAG CAG CGA ACT TCA CA 3’ | |
| qnrB-R | 5’ GAT GCC AAG CCG CTC CAT GA 3’ | |
| qnrS | 219 | |
| qnrS-F | 5’ TCA CCT TCA CCG CTT GCA CAT T 3’ | |
| qnrS-R | 5’ AAT CAC ACG CAC GGA ACT CTA TAC C 3’ | |
| gyrA | 360 | |
| gyrA-F | 5’ GCT GCC AGA TGT CCG AGA T 3’ | |
| gyrA-R | 5’ TCC GTG CCG TCA TAG TTA TCA 3’ |
3.6. Amplification of gyrA Gene by PCR
The gyrA gene was amplified using specific primers (Table 1). PCR reactions were performed in a final volume of 25 µL containing 10 pmol of each primer, 1 µg of extracted DNA, 200 µM of nucleoside triphosphate, and one unit of Taq DNA polymerase. Amplification was performed with pre-incubation (94ºC for five minutes), followed by 40 cycles of denaturation (94ºC for 30 seconds), annealing (63ºC for 30 seconds), extension (72ºC for 20 seconds), and a final extension (72ºC for 10 minutes). PCR products of qnr and gyrA genes were electrophoresed on agarose gel containing DNA safe stain. DNAs were visualized under UV light, and band sizes were measured using size marker.
3.7. PCR-RFLP
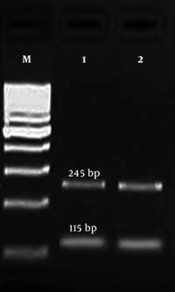
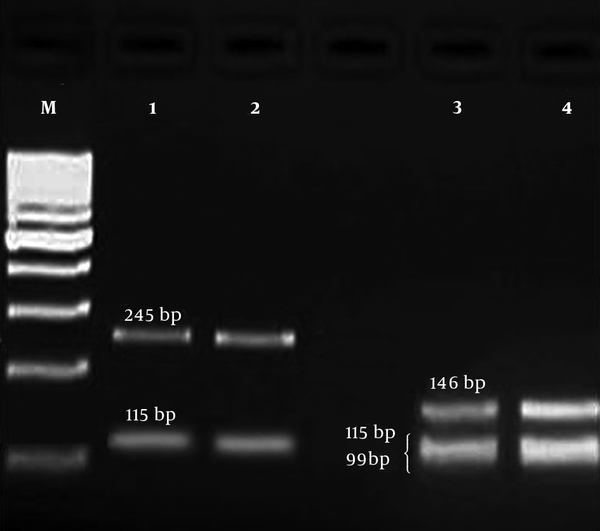
PCR product of gyrA gene was digested by HinfI enzyme to identify the mutation in gyrA gene by restriction fragment length polymorphism (RFLP) method. A point mutation in gyrA gene at ser-83 (T → G) is related to resistance to fluoroquinolones. In wild types, there are two restriction sites for HinfI, and after digestion, three fragments (99, 115, and 146 bp) are produced (13). But, in mutant strains, one restriction site is deleted and only two fragments (115 and 245 bp) are observed (Figure 1).
Agarose gel electrophoresis of PCR-RFLP products for gyrA gene; M: 100-bp DNA size marker, columns 1 and 2: mutant isolates (produced two fragments, 115 and 245 bp), columns 3 and 4: wild type isolates (produced three fragments, 99, 115, and 146 bp).
3.8. Sequencing
In order to confirm whether the observed bands were related to those genes, PCR products were sent to the Bioneer Company in South Korea for sequencing and sequences were compared using online NCBI/BLAST software.
3.9. Statistical Analysis
The analysis was performed using SPSS version 21.0 (IBM Corp., USA). Chi-square or the Fisher exact tests were used to determine the significance of differences. The difference was considered statistically significant if the P value was less than 0.05.
4. Results
4.1. Antibiotic Resistance Pattern
Totally, 309 E. coli isolates were collected from 1723 clinical samples. Out of the 309 isolates, the highest resistance rate was against nalidixic acid (60.2%), and the lowest rate was against norfloxacin (40.5%). The results of antibiotic susceptibility testing are shown in Table 2.
| Antibiotic | Susceptible | Intermediate | Resistant |
|---|---|---|---|
| Norfloxacin | 58.6 | 0.9 | 40.5 |
| Ofloxacin | 55.7 | 0.6 | 43.7 |
| Norfloxacin | 36.6 | 3.2 | 60.2 |
4.2. Distribution of Quinolones Resistance Genes
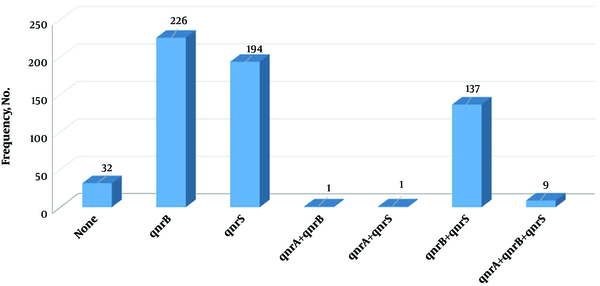
All the clinical isolates were examined for the presence of qnrS, qnrB, and qnrA genes. The highest frequency of qnr genes belonged to qnrB (73.1%). Moreover, 32 isolates (10.4%) had no qnr genes, and nine strains (2.9%) contained all three genes (Figure 2).
Frequency of simultaneous presence of qnr genes
4.3. Association of Antibiotic Resistance and qnr Ggenes
In the present study, the association between the presence of those three genes with resistance to three quinolones was detected. First of all, the association between the presence of qnrA gene and resistance to nalidixic acid, norfloxacin, and ofloxacin among the susceptible and resistant isolates was examined, and there was a significant relationship only between the presence of qnrA gene and the resistance to norfloxacin (P = 0.016). Then, the association between the presence of qnrB gene and resistance to the same antibiotics among the susceptible and resistant isolates was investigated, and a significant relationship was only observed between the presence of this gene and resistance to nalidixic acid (P = 0.037). Also, the study on the association between the presence of qnrS gene and the examined antibiotics revealed no significant relationship between the presence of qnrS gene and resistance to those three antibiotics.
Based on the present study results, eight isolates resistant to norfloxacin contained all the three qnrS, qnrB, and qnrA genes. Meanwhile, the simultaneous presence of qnrB and qnrS genes with the frequency of 46.4% showed the highest rate of the co-existence of these genes in the resistant isolates. Only seven isolates (3.8%) out of the total isolates resistant to nalidixic acid included all the three genes, and 85 isolates (45.7%) included qnrS and qnrB. However, seven isolates resistant to ofloxacin included all the three genes. Among the isolates resistant to this antibiotic, the highest rate of simultaneous presence of genes was related to qnrS and qnrB among 59 strains (43.7%).
4.4. Distribution of gyrA Mutation
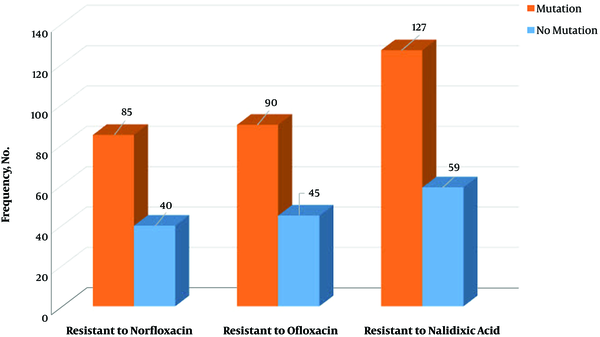
On the other hand, among 309 tested E. coli strains, a mutation in gyrA gene was observed in 174 (56.3%) isolates. The mutation was observed in codon 83, which conferred resistance to fluoroquinolones. Also, there was a significant relationship between resistance to the tested antibiotics and mutation in gyrA gene (P = 0.01). Meanwhile, in 39 isolates with gyrA mutation, no antibiotics resistance was observed (Figure 2). Finally, there were no significant relationships between the presence of qnr genes and mutation in gyrA gene (P = 0.538) (Figure 3).
Correlation of antibiotic resistance and mutation in gyrA gene at codon 83
Correlation of qnr genes and mutation in gyrA gene at codon 83
5. Discussion
Nowadays, quinolones are frequently used to treat UTIs. Multi-drug resistance microorganisms, particularly extended spectrum beta-lactamases (ESBLs), producing ones, increased in Gram-negative bacteria as a result of the misuse of antibiotics (14-17). The present study aimed at investigating the frequency of quinolone resistance in E. coli strains isolated from patients referred to hospitals in the North of Iran. Out of 1723 samples, 309 E. coli strains were isolated, among which 186 isolates (60.2%) were resistant to nalidixic acid. The result of this study was similar to that of Mansory Jamshidi et al. in Tehran, Iran (18); but resistance rate to nalidixic acid in the current study was higher than those of Alos et al. (19), and Moreno et al. in the United States (20), as well as Alshara in Jordan (21) and Nakhaei Moghadam et al. in Mashhad, Iran (22). The difference between the findings of the study in the United States and those of the current study regarding the resistance rate could be due to supervised drug consumption program and no access to non-prescribed drugs in that country. Although most clinical strains of E. coli in the United States and Canada are susceptible to fluoroquinolones, the strains resistant to these antibiotics are increasing (19).
In the present study, in addition to nalidixic acid, the resistance to two fluoroquinolones such as norfloxacin and ofloxacin was investigated. In this regard, 125 isolates (40.5%) resistant to norfloxacin and 135 isolates (43.7%) resistant to ofloxacin were observed, and norfloxacin was the most effective antibiotic against E. coli isolates in the present study. Goettsch et al. in the Netherlands studied resistance to the antibiotics most commonly prescribed to treat UTIs. From 1989 to 1998, in more than 90,000 E. coli isolates, resistance to norfloxacin increased from 1.3% in 1989 to 5.8% in 1998. Niranjan et al. in Northern India worked on 119 E. coli isolates, out of which 74.2% were resistant to norfloxacin. The reported rate was higher than that of the current study, and resistance to ofloxacin was not investigated in their study (23). Determination of the resistance rate to quinolones is very important since they are frequently used as the drug of choice to treat infections caused by E. coli; and, therefore, inappropriate treatment with these antibiotics can lead to treatment failure.
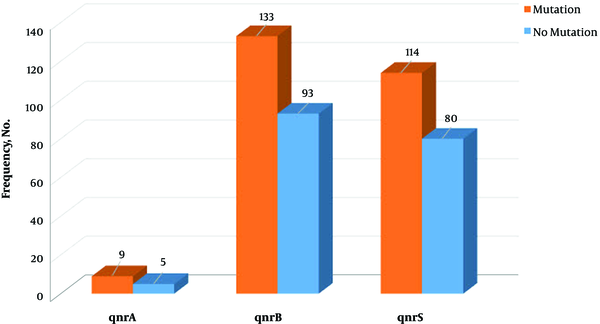
One of the mechanisms of bacterial resistance to fluoroquinolones is plasmid-mediated resistance or resistance related to the presence of qnr gene. In the present study, out of the total 309 clinical examined isolates, 14 (4.5%) were positive for qnrA, 226 (73.1%) for qnrB, and 194 (62.8%) for qnrS genes. Meanwhile, 32 samples (10.4%) showed none of the qnr genes. Castanheira et al. in Brazil, among 144 E. coli isolates, reported one (0.69%) qnrA-producing isolate. It was the first report of a qnrA-carrying isolate in a Latin American country, which was lower than the result of this study. However, no frequency of qnrB and qnrS was detected (24). In a study in Morocco by Bouchakour et al. the prevalence of qnrA, qnrB, and qnrS genes was 2.56%, 10.25%, and 23.07, respectively. Mansory Jamshidi et al. examined the prevalence of qnr genes in 150 E. coli isolates. The prevalence of qnrA, qnrB, and qnrS were reported 31.8%, 56.5%, and 28.9% respectively, and only the prevalence of qnrA was higher than those of the current study; the prevalence of qnrB and qnrS were not reported (18).
The present study also investigated the relationship between the presence of qnr genes and the resistance to antibiotics. The association between the presence of qnrA gene and resistance to nalidixic acid, norfloxacin, and ofloxacin among the isolated bacteria was examined, and there was only a significant association between the presence of qnrA and the resistance to norfloxacin. Furthermore, the relationship between the presence of qnrB gene and the resistance to the three antibiotics among the isolates was examined, and a significant association was observed between the presence of this gene and resistance to nalidixic acid, and no significant association was observed between qnrS and resistance to the antibiotics. Also, there was a significant relationship between a mutation in gyrA gene and resistance to the three antibiotics.
5.1. Conclusions
In summary, our findings pointed out the prevalence of quinolones resistance and the most prevalent resistance mechanisms among clinical isolates of E. coli in the North of Iran. Moreover, despite the significant association of the investigated genes with fluoroquinolones resistance in the current study tested isolates, the presence of these genes in susceptible strains suggested that some other possible mechanisms could influence fluoroquinolones resistance.
Acknowledgements
References
-
1.
Al-Badr A, Al-Shaikh G. Recurrent urinary tract infections management in women: a review. Sultan Qaboos Univ Med J. 2013;13(3):359-67. [PubMed ID: 23984019]. [PubMed Central ID: PMC3749018].
-
2.
Ebrahim-Saraie HS, Nezhad NZ, Heidari H, Motamedifar A, Motamedifar M. Detection of antimicrobial susceptibility and Integrons among extended-spectrum beta-lactamase producing uropathogenic Escherichia coli isolates in Southwestern Iran. Oman Med J. 2018;33(3):218-23. [PubMed ID: 29896329]. [PubMed Central ID: PMC5971052]. https://doi.org/10.5001/omj.2018.40.
-
3.
Asgharzadeh Kangachar S, Mojtahedi A. The presence of extended-spectrum β-lactamase as a risk factor for MDR in clinical isolation of Escherichia coli. Trop Biomed. 2017;34(1):98-109.
-
4.
Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol. 2015;13(5):269-84. [PubMed ID: 25853778]. [PubMed Central ID: PMC4457377]. https://doi.org/10.1038/nrmicro3432.
-
5.
Bennett PM. Plasmid encoded antibiotic resistance: acquisition and transfer of antibiotic resistance genes in bacteria. Br J Pharmacol. 2008;153 Suppl 1:S347-57. [PubMed ID: 18193080]. [PubMed Central ID: PMC2268074]. https://doi.org/10.1038/sj.bjp.0707607.
-
6.
Malekzadegan Y, Hadadi M, Ebrahim-Saraie HS, Heidari H, Motamedifar M. Antimicrobial resistance pattern and frequency of multiple-drug resistant Enterobacter spp. at a tertiary care hospital in Southwest of Iran. J Krishna Inst Med Sci. 2017;6(2):33-9.
-
7.
Aldred KJ, Kerns RJ, Osheroff N. Mechanism of quinolone action and resistance. Biochemistry. 2014;53(10):1565-74. [PubMed ID: 24576155]. [PubMed Central ID: PMC3985860]. https://doi.org/10.1021/bi5000564.
-
8.
Heisig P. Genetic evidence for a role of parC mutations in development of high-level fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 1996;40(4):879-85. [PubMed ID: 8849244]. [PubMed Central ID: PMC163223].
-
9.
Fu Y, Zhang W, Wang H, Zhao S, Chen Y, Meng F, et al. Specific patterns of gyrA mutations determine the resistance difference to ciprofloxacin and levofloxacin in Klebsiella pneumoniae and Escherichia coli. BMC Infect Dis. 2013;13:8. [PubMed ID: 23295059]. [PubMed Central ID: PMC3576228]. https://doi.org/10.1186/1471-2334-13-8.
-
10.
Tayebi Z, Heidari H, Kazemian H, Ghafoori SM, Boroumandi S, Houri H. Comparison of quinolone and beta-lactam resistance among Escherichia coli strains isolated from urinary tract infections. Infez Med. 2016;24(4):326-30. [PubMed ID: 28011969].
-
11.
Asgharzadeh Kangachar S, Mojtahedi A. Evaluation of integrons classes 1–3 in extended spectrum beta-lactamases and multi drug resistant Escherichia coli isolates in the North of Iran. Gene Reports. 2018;12:299-303. https://doi.org/10.1016/j.genrep.2018.07.014.
-
12.
Wayne P. Performance standards for antimicrobial susceptibility testing. 26th ed. Clinical and Laboratory Standards Institute (CLSI); 2016.
-
13.
Ruiz J. Mechanisms of resistance to quinolones: target alterations, decreased accumulation and DNA gyrase protection. J Antimicrob Chemother. 2003;51(5):1109-17. [PubMed ID: 12697644]. https://doi.org/10.1093/jac/dkg222.
-
14.
Soltani B, Heidari H, Ebrahim-Saraie HS, Hadi N, Mardaneh J, Motamedifar M. Molecular characteristics of multiple and extensive drug-resistant Acinetobacter baumannii isolates obtained from hospitalized patients in Southwestern Iran. Infez Med. 2018;26(1):67-76. [PubMed ID: 29525800].
-
15.
Tavakoly T, Jamali S, Mojtahedi A, Khan Mirzaei M, Shenagari M. The prevalence of CMY-2, OXA-48 and KPC-2 genes in clinical isolates of Klebsiella spp. Cell Mol Biol (Noisy-le-grand). 2018;64(3):40-4. [PubMed ID: 29506629]. https://doi.org/10.14715/cmb/2018.64.3.7.
-
16.
Haghighatpanah M, Mozaffari Nejad AS, Mojtahedi A, Amirmozafari N, Zeighami H. Detection of extended-spectrum beta-lactamase (ESBL) and plasmid-borne blaCTX-M and blaTEM genes among clinical strains of Escherichia coli isolated from patients in the north of Iran. J Glob Antimicrob Resist. 2016;7:110-3. [PubMed ID: 27721192]. https://doi.org/10.1016/j.jgar.2016.08.005.
-
17.
Ghaffarian F, Hedayati M, Sedigh Ebrahim-Saraie H, Atrkar Roushan Z, Mojtahedi A. Molecular epidemiology of ESBL-producing Klebsiella pneumoniae isolates in intensive care units of a tertiary care hospital, North of Iran. Cell Mol Biol (Noisy-le-grand). 2018;64(7):75-9. [PubMed ID: 29974849].
-
18.
Mansory Jamshidi N, Pakzad E, Tabaraee B, Hadadi A. Frequency of qnr genes in Escherichia coli strains resistant to quinolones isolated from Ilam Imam khomani hospital and Tehran Milad hospital. J Ilam Univ Med Sci. 2013;21(6):16-22.
-
19.
Alos JI, Serrano MG, Gomez-Garces JL, Perianes J. Antibiotic resistance of Escherichia coli from community-acquired urinary tract infections in relation to demographic and clinical data. Clin Microbiol Infect. 2005;11(3):199-203. [PubMed ID: 15715717]. https://doi.org/10.1111/j.1469-0691.2004.01057.x.
-
20.
Moreno E, Prats G, Sabate M, Perez T, Johnson JR, Andreu A. Quinolone, fluoroquinolone and trimethoprim/sulfamethoxazole resistance in relation to virulence determinants and phylogenetic background among uropathogenic Escherichia coli. J Antimicrob Chemother. 2006;57(2):204-11. [PubMed ID: 16390858]. https://doi.org/10.1093/jac/dki468.
-
21.
Alshara M. Antimicrobial resistant pattern of Escherichia coli strains isolated from pediatric patients in Jordan. Acta Med Iran. 2011;49(5):293-5. [PubMed ID: 21713746].
-
22.
Nakhaei Moghadam M, Moshrefi S. Determining the antibiotic resistance pattern of urinary isolates of Escherichia coli and prevalence of extended spectrum β-lactamases (ESBLs) among them. J Sabzevar Univ Med Sci. 2010;16(4):228-33.
-
23.
Niranjan V, Malini A. Antimicrobial resistance pattern in Escherichia coli causing urinary tract infection among inpatients. Indian J Med Res. 2014;139(6):945-8. [PubMed ID: 25109731]. [PubMed Central ID: PMC4165009].
-
24.
Castanheira M, Pereira AS, Nicoletti AG, Pignatari AC, Barth AL, Gales AC. First report of plasmid-mediated qnrA1 in a ciprofloxacin-resistant Escherichia coli strain in Latin America. Antimicrob Agents Chemother. 2007;51(4):1527-9. [PubMed ID: 17220425]. [PubMed Central ID: PMC1855464]. https://doi.org/10.1128/AAC.00780-06.