Abstract
Background:
Helicobacter pylori bacterium is responsible for gastritis, peptic ulcer, gastric cancer, duodenal ulcer, and mucosal membrane lymphoma. There is a tremendous variety among H. pylori strains. By 2010, ten different populations of H. pylori were identified. The geographical differences between strains may be the explanation for the difference in prevalence of diseases associated with H. pylori in some areas. Various methods have been developed, such as PFGE Southern Blot Analysis-Ribotyping Plasmid Analysis, Rep-PCR and multilocus sequence typing (MLST) for typing H. pylori.Objectives:
This study was conducted to investigate the molecular epidemiology and gene variation of H. pylori isolates from patients with gastroduodenal diseases using sequence typing of seventy structural genes.Methods:
Two biopsy samples were taken from patients with gastrointestinal disorders. The urease test was performed on one of the samples, the other was cultured on Columbia agar with sheep blood, and then DNA was extracted. Seven structural genes were proliferated with designed primers and then were sequenced. The results were recorded on the MLST and Genbank, as well as the allele numbers and sequence types were determined. Data were analyzed by MEGAv6 and START v2 software.Results:
The allele numbers and sequence types of H. pylori strains were unique, and were not recorded previously in the MLST database. The strains of H. pylori isolated from Kermanshah belonged to the hpEurop population.Conclusions:
Due to the unique nature of strains and their various sequences, the strains were located in separate clones. Accordingly, none of them could handle the role of type strain in Kermanshah.Keywords
1. Background
Helicobacter pylori is a Gram-negative, motile, spiral-shaped bacterium colonized in the human stomach mucosa (1). It is a bacterium causing chronic gastritis, gastric ulcer and duodenal ulcer, as well as a risk factor for stomach cancer, mucosa-associated lymphoma and gastric adenocarcinoma (2-5). The mechanism of H. pylori transmission has not yet fully disclosed, whose reservoir is the human stomach and transmission may occur through mother-to-child transmission (vertically) or individual transmission (horizontally). The main transmission routes in families are either person-to-person via oral-oral or oral-fecal or environmental contact, and through water in developing countries (6).
The H. pylori infection is usually acquired, and if antibiotics do not root it out, it will sustain for decades or throughout life (7, 8). The prevalence of H. pylori infection in different geographical regions is very diverse. The prevalence of H. pylori infection is 25% in developed countries and more than 80% in developing countries (9). The H. pylori that have been distributed globally infect more than half of the world’s population and it is important in the general health of the community, but most people infected with this bacterium are asymptomatic, and the infection remains in its latent state (10). Only 15% - 20% of infected people develop gastroduodenal diseases. One possible reason is that H. pylori has high genetic diversity and intra-species variation and the patients are infected with different H. pylori strains (11).
The genetic diversity in the H. pylori population is greater than the population of other bacteria (12) and is about 50 times higher than the human population (5, 13, 14) This high diversity among populations is a description of high mutation and recombination rates in H. pylori strains (12, 15). Different phylo-geographic strains of H. pylori isolated from the gastric biopsy of patients with gastroduodenal diseases in different geographical regions indicate that this bacterium is diverse in the ethnic and geographical structure (12, 16). Despite the high incidence of H. pylori infection in Africa and South Asia, the prevalence of gastric cancer in these regions is much lower than in other countries and the prevalence of gastric cancer is reduced from north to southeast Asia. In addition to host factors, the environmental and nutritional factors as possible causes for different outcomes of H. pylori infection are related to the differences in the pathogenicity of H. pylori strains, and geographical differences can be an explanation for the difference in pathology.
There is a high geographical diversity in Iran, and nearly 69% of the Iranian population is infected with H. pylori (17, 18). Instead, a small number of clones are more prevalent among a specific population (19). The geographical difference in H. pylori strains has been shown by various studies, including the use of molecular typing methods. Based on the geographical relationship and using the multilocus sequence typing (MLST) method by 2010, seven modern types of populations of H. pylori (hpEurope, hpEastAsia, hpAfrica1, hpAfrica2, hpAsia2, hpNEAfrica and hpSahul) and three subtypes of H. pylori (hspMaori, hspAmerind, hspEAsia) have been identified (20, 21).
The MLST method was suggested in 1998 as a tool for studying the epidemiology and typing of bacteria. Genetic characteristics of the strains are examined by using the MLST method and the ancestral relation between isolates is recognized by analyzing the geographical origin of the strains. In this method, applied information is obtained from the genetic variation of pathogenic bacteria based on the DNA sequence of several structural genes. This information for their epidemiological studies is appropriate and reflects the biology and evolution of the population (22). In the global epidemiology of pathogenic strains in a certain geographic region, their association is determined with other strains isolated from different parts of the world. In the MLST method, the file of sequencing data is compared between different laboratories and the data saved in the central database is accessible through the Internet and provides a potential source for the study of global epidemiology (23-25).
2. Objectives
The aim of this study was to analyze the sequence of structural genes in order to identify genetic diversity patterns and new sequence types, to investigate the clonal relationships between strains isolated from Kermanshah and to determine the strain type in isolated clones.
3. Methods
3.1. Study Subjects
The present descriptive-analytical and cross-sectional study was conducted on 30 strains of H. pylori during 2014 to 2015 isolated from gastric biopsy cultures of patients referred to Endoscopy Center of Imam Khomeini Hospital in Kermanshah (Iran). The patients with symptoms of gastric pain, gastric ulcer, reflux, dysphasia, indigestion, and atrophy were included in this study. The patients who were taking the anti-inflammatory, steroidal and anti-acid drugs or antibiotics two weeks prior to sampling were also excluded from the study, as well as patients with bleeding in the stomach. Informed consent was recorded for each patient who experienced endoscopy under the supervision of a specialist physician. Clinical findings and patient profile were recorded for each patient.
3.2. Isolation and Identification of Bacteria
A gastroenterologist prepared two biopsy samples of stomach corpus and antrum from each patient. A rapid urease test was performed for one of the samples. The biopsy samples were placed in 1 mL of transport medium of brain heart infusion broth with 30% glycerol. The samples were transferred under a cold chain condition to the laboratory at the School of Medicine in 4 hours, the biopsy samples were cultured on Brucella agar selective medium enriched with 10% defibrinated sheep blood, and 10% decomplemented fetal bovine serum. The plates were incubated under microaerophilic conditions in the presence of 5% O2, 15% CO2, 80% N2 and 90% humidity for 3 to 7 days at 37°C. Bacterial colonies were identified based on colony morphology, Gram staining, and bacterial spiral shape under the microscope, as well as positive urease, catalase and oxidase tests.
3.3. DNA Extraction, PCR
After cultivating the biopsy specimens, isolated bacteria were cultured on a culture medium using a swab in a microtip containing a physiological serum and then homogenized with vertex and isolated on the sediment from the bacterium. DNA extraction was performed according to Sina kit’s instruction clone. Also, PCR reaction in total volume of 30 µL with the final concentration of PCR master mix (1×) and the final concentration of forward primer (0.5 µM), and the final concentration of primer (reverse (0.5 µM) extracted DNA concentration of 100 ng was done.
On the DNA extracted from each isolate, the replication of seven structural genes (atsA, efp, mutY, ppa, trpC, ureI, vacA and yphC) was done using PCR and primers and the annealingtemperature listed in the table by the c1000-thermal cycler Biorad apparatus sn: 3030004957, Singapore and a positive control sample and a negative control sample were generated in each PCR series. The PCR amplification electrophoresis was performed on a 1% agarose gel, which appeared with ethidium bromide, and then the gel was examined in the xr + gel documentation (sn; 721br02272, USA) using a Quantity-One-Installer software.
3.4. Determination of the Sequence of the Gene and Mlst
Each PCR product was amplified and sequenced by sanger method (Bioneer, South Korea) using forward and reverse primers. All chromatograms were analysed and edited using Bioedit software. For achieving full length of each locus, edited sequence of forward primer and edited sequence of complementary reverse of reverse primer were paired. All sequences generated in this study were recorded in the Genbank database, along with their translated proteins. The accession numbers for the registered genes are as follows:
- KT198757-KT198786(atpA)
- KT198787-KT198816(efp)
- KT198817-KT198846(mutY)
- KT198847-KT198876(ppa)
- KT198877-KT198906(trpC)
- KT198907-KT198936(ureI)
- KT198937-KT198966(yphC)
Then, the sequences were recorded on the MLST website, determining the allele number assigned to each locus. The allele numbers assigned to each of the seven genes for each isolate were recorded in the MLST profile, and the sequence types for each isolate were determined. The allele numbers and sequence types were compared with the data on the MLST database. An allelic profile of seven integers and an exclusive allelic profile for the sequence types were specified for each isolate. The START (sequence type analysis and recombinational tests) was used as a selected tool useful in the analysis of sequence types and MLST data. The index of association was determined to measure the linkage disequilibrium between the alleles in the START program. Finally, MEGA (molecular evolutionary genetics analysis) software was used to study the similarity of phylogenetic tree of strains according to Neighbor- joining method.
3.5. Data Analysis
Version 21 of SPSS software was employed for the statistical analysis. The data were analyzed using chi-square. The P < 0.05 was considered statistically significant.
4. Results
The age range of patients was between 20 to 76 years with an average age of 48.6 years, including 13 (43.3%) females and 17 (56.7%) males. Based on the diagnosis of a specialist, 18 (60%) patients had gastritis and 12(40%) cases showed duodenal ulcer. All aspects of the H. pylori isolates including antibiotic resistance have been studied and published in another article (26).
4.1. DNA Extraction and PCR
The integers of seven loci for 23 isolates from H. pylori were proliferated and sequenced. Each of the seven structural genes (atpA, efp, mutY, ppa, trpC, ureI, yphC) were proliferated using primers previously designed and described Table 1. The PCR product was observed within the bands of 834; 599; 661; 706; 633; 636; 734 bp. The quality and purity of the PCR products were confirmed by electrophoresis on gel (Figure 1).
The Primers Used and the Size of the Reproduced Parts of the Genes
| Gene/Primer | Amplification and Sequencing | Size of PCR Products | Annealing Temperature |
|---|---|---|---|
| atpA | 842 | 61°C, 30 s | |
| F | GGACTAGCGTTAAACGCA’CG | ||
| R | CTTGAAACCGACAAGCCCAC | ||
| efp | 559 | 58°C, 30 s | |
| F | GGCAATTTGGATGAGCGAGCTC | ||
| R | CTTCACCTTTTCAAGATACTC | ||
| mutY | 661 | 54°C, 30 s | |
| F | GTGGTTGTAGYTGGAAACTTTACAC | ||
| R | CTTAAGCGTGTGTYTTTCTAGG | ||
| ppa | 706 | 51°C, 30 s | |
| F | GGAGATTGCAATGAATTTAGA | ||
| R | GTGGGGTTAARATCGTTAAATTG | ||
| trpC | 633 | 61°C, 30 s | |
| F | TAGAATGCAAAAAAGCATCGCCCTC | ||
| R | TAAGCCCGCACACTTTATTTTCGCC | ||
| ureI | 686 | 51°C, 30 s | |
| F | AGGTTATTCGTAAGGTGCG | ||
| R | GTTTAAATCCCTTAGATTGCC | ||
| yphC | 734 | 54°C, 30 s | |
| F | CACGCCTATTTTTTTGACTAaAAAAC | ||
| R | CATTYACCCTCCCAATGATGC |
Electrophoresis of PCR product of seven structural genes of Helicobacter pylori

The sequence of seven structural gene segments are compared on the MLST and the characteristics of isolates are checked with central databases via the Internet, while the size of DNA fragments on the gel is compared in other typing methods (27). The allelic profile and sequence types are obtained by using the MLST of a combination of seven structural gene alleles. The relationship between isolates is achieved by plotting dendrogram of pairwise matrix between different sequence types. In the MLST method, the alleles are identified in structural genes directly by sequencing the nucleotides (28).
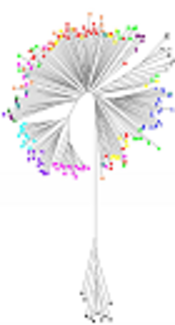
Table 2 shows that each of the 23 sequence types are unique and H. pylori not already described on the MLST database. Two pairs of strains had a common sequence type, as a pair for unrelated patients with biopsy obtained on the same day, another pair for unrelated patients with biopsy achieved within a week. Most allele numbers derived from 23 strains are unique, except for two pairs of strains that had common allele numbers for all loci. A phylogenic tree was drawn using sequences classified in seven structural genes. This tree provided evidence that isolated strains are clustered into three groups (Figure 2).
The Sequence Types of Isolates
| Sequence Type | Disease | Age, y | Sex | ID |
|---|---|---|---|---|
| ST-2906 | Gastritis | 61 | Female | 1Ke |
| ST-2907 | Gastritis | 57 | Male | 2Ke |
| ST-2908 | Gastritis | 48 | Female | 3Ke |
| ST-2909 | Peptic ulcer | 48 | Male | 4Ke |
| ST-2910 | Peptic ulcer | 50 | Female | 5Ke |
| ST-2911 | Gastritis | 50 | Male | 6Ke |
| ST-2912 | Gastritis | 30 | Male | 7Ke |
| ST-2913 | Gastritis | 54 | Male | 8Ke |
| ST-2914 | Peptic ulcer | 76 | Female | 9Ke |
| ST-2915 | Gastritis | 55 | Female | 10Ke |
| ST-2916 | Gastritis | 28 | Male | 11Ke |
| ST-2917 | Gastritis | 20 | Female | 12Ke |
| ST-2918 | Gastritis | 26 | Female | 13Ke |
| ST-2917 | Peptic ulcer | 38 | Male | 14Ke |
| ST-2920 | Gastritis | 36 | Male | 15Ke |
| ST-2921 | Peptic ulcer | 68 | Male | 16Ke |
| ST-2922 | Peptic ulcer | 40 | Male | 17Ke |
| ST-2923 | Gastritis | 70 | Male | 18Ke |
| ST-2924 | Gastritis | 50 | Male | 19Ke |
| ST-2924 | Peptic ulcer | 36 | Male | 20Ke |
| ST-2926 | Gastritis | 27 | Male | 21Ke |
| ST-2927 | Gastritis | 61 | Female | 22Ke |
| ST-2931 | Peptic ulcer | 38 | Female | 26Ke |
Evolutionary relationship between Helicobacter pylori strains isolated from Kermanshah in Iran; evolutionary history inferred from the sequence of seven structural genes of H. pylori isolates using the “neighbor-joining” method

The isolates, sequence types and allele numbers were inserted into the STARTv2 software as a useful tool for analyzing MLST data, and then were analyzed using the UPGAMA (unweighted pair group mean average) tool. Cluster analysis and distance matrix indicate that the isolated strains are clustered into three groups; and the calculated distance indicates that 19Ke isolate with 20Ke isolate as well as 12Ke isolate with 14Ke isolate are genetically identical. The 5Ke isolate is closely related to 26Ke isolated (Figure 3).
Dendrogram plotted by START2 software to analyze the relationship by UPGAMA

4.2. Calculate the Index of Association in the START Software
This statistical test is used to measure the balance of intra-population relationships, the amount of recombination among a set of sequences, and the recognition of association between alleles in different locations. Neisseria gonorrhoeae has a panmictic structure with random association between alleles. Many bacterial strains such as Salmonella, a non-random association of alleles, have a strong association between alleles in terms of clonal population structure, which has no high chromosomal recombination rates to break the clonal association. In the past, the association of special serotypes with diseases suggested the clonal nature of the bacteria, while the allele-specific alteration index in several chromosomes is very successful in generating a large number of data for statistical analysis of bacterial populations.
The clonal nature has the dominant mutation, and the genetic diversity is produced, but there is also a significant association, significant association=linkage disequilibrium (19). The clonal MLST groups are defined by having multiple identical allele numbers. In the clonal H. pylori populations, the mutation is more prevalent in genetic variation and a significant association exists in alleles in different loci. In the non-clonal populations, while the recombination is more dominant, the lower degree of association can be seen between the different loci. The association between the alleles in the structural genes is calculated by the index of association. Genetic diversity occurs in H. pylori by recombination or mutation.
In the present study, IA = (Vo / Ve) - 1 = the index of association was obtained to be 3.577 for all isolates; there is no evidence for association. The isolates (1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 13, 15, 16, 17, 18, 21 and 22), which have no same allele number, showed the index of association of -0.003. When the expected value of the index of the association is zero, there will be no association between the alleles, indicating a non-clonal bacterial population, a predominant recombination, and a lower association between the alleles. The IA was 6.128 for 5, 12, 14, 19, 20 and 26 isolates, when the index of association is distinctly different from zero. This indicates that there is little or no recombination when clonal structure of population is found at all levels, so the index of association is greater than zero, mutation is more prevalent in generating genetic variation, and the association between alleles is greater.
4.3. Multilocus Sequence Analysis
All strains of H. pylori from Kermanshah have homology similar to those of the hpEurope sub-population, generally, by comparing 23 MLST profiles based on DNA sequencing from seven structural genes classified by Kermanshah isolates with genomic analysis of 174 H. pylori sequences of different geographical regions and ethnic groups different from other populations (Africa, Europe, Australia and Asia) and other Iranian isolates (Table 3). The neighbor-joining phylogenetic tree analysis was performed with the MEGA software (Figure 4), which clearly revealed the geographical distribution of the diverse population of H. pylori and related sub-populations. This analysis showed the integration of all strains isolated from Kermanshah and other Iranian isolates close to the population of Spain-Estonia belonging to the population hpEurope.
Selected Strains for Comparison with Helicobacter pylori Strains Isolated from Kermanshah
| Country | hpAfrica1 | hpNEAfrica | hpEurope | hpEAsia | hpAfrica2 | hpmaoril | hpAsia2 | hpAmerind |
|---|---|---|---|---|---|---|---|---|
| Ethiopia | 19 | |||||||
| Angola (South Africa) | 15 | 10 | ||||||
| Spain | 20 | |||||||
| Finland | 9 | |||||||
| Estonia | 11 | |||||||
| UK | 9 | |||||||
| Italy | 6 | |||||||
| Malaysia | 1 | |||||||
| Thailand | 1 | |||||||
| Chine | 6 | |||||||
| Philippine | 2 | |||||||
| Kazakhstan | 1 | |||||||
| USA | 7 | |||||||
| New Zealand | 15 | |||||||
| Japan | 3 | |||||||
| Colombia | 4 | |||||||
| Iranian | 35 |
The neighbor-joining tree of the population structure of a group of Helicobacter pylori, while the H. pylori strains of Kermanshah are yellow

5. Discussion
The isolates of H. pylori from Kermanshah were tested and evaluated. The MLST method is a typing system reflecting the population and the evolutionary biology of bacteria and provides the ability to compare the results between different laboratories. Although the number of isolates is relatively small, the diversity of isolates has been proven by identifying the unique allele numbers and sequence types. Most sequence types are new, and have never been found in the MLST database for H. pylori (29). Countries with populations from multiple regions provide a good opportunity to study further the H. pylori population structure (9).
Iran with a developed agriculture and civilization is an Islamic country, is situated at the intersection of Europe, Asia, and Africa. It is a vast country with diverse ethnic groups (a combination of ethnicities including Kurd, Lur, Fars, Gilaki, Arab, and Azeri ethnos). Ethnic and geographical diversity in Iran is the reflection of the historical interaction with other populations. The Arab population of Iran entered the country in the seventh and eighth century during the Islamic victory over the Iranian Empire. The Kurd population in northwestern Iran reveals a historical connection with Turkish Kurds and other Turks during several eras of history, including the Ottoman Empire by 1514, during the First World War and in parts of the 20th century (16).
It is estimated that 69% of the Iranian population currently suffer from H. pylori infection. It has been shown that the frequency of gastrointestinal ulcer and gastric cancer is largely influenced by geographical conditions and ethnic groups (16). Iran has had a mixed population of diverse ethnic groups throughout history. Therefore, the chances of DNA transmission between strains of different genotypes may be higher when compared to other countries (16). The prevalence of gastric cancer in Iran is high, indicating significant geographical differences among H. pylori strains. Kermanshah is a city with a 947,000 population, with Kurd ethnicity located in northwestern Iran. People eat meals together, increasing the risk of H. pylori infection, and leading to more genetic variation in this bacterium. The genetic analysis of the population by MLST showed that H. pylori has no clonal structure, but has a panmictic structure. Horizontal gene transfer and frequent recombination lead to extensive genetic rearrangement. Helicobacter pylori seems to have a clonal structure over a short period; for example, when shifting from one host to another (30).
It seems that H. pylori and humans have a historical connection. In addition, direct evidence has been observed for recombination between the different strains of H. pylori in humans and mice. However, several lines of evidence have shown that the mutation may play an important role in generating the genetic variation observed in H. pylori. There are documents on recognition of the clonal generation in some family groups in Germany and Japan. The mutation is more dominant in the clonal structure of the H. pylori population (29). In line with our study, Falush et al. suggested that the H. pylori populations have no clonal structure, probably due to diverse recombination between heterozygous strains during mixed colonization. The results of this study indicate that most alleles are associated with unrelated H. pylori strains because there is a high level of recombination among H. pylori isolates. As a result, most isolates appear to have different sequence types though the recombination may occur for most of the genes studied on a group scale.
Two complexes of poor clonal structures are considered, while it has been proposed that more clonal classes would be identified if more samples were taken. It is true that the clonal structure of H. pylori population is slowly declined by recombination. The phylogenetic tree generated from the allelic profile provides evolutionary information on isolates regarding UPGMA and analytical analysis of allelic profile data through the H. pylori genome indicates a high degree of genetic variation among strains. The clonal groups are known to be weak, while placed on a free recombination of patterns, providing a result based on further analysis with population genetics. The index of association was 3.577 in the current study and 5.27 in a study conducted by Liao et al. (29) These results, consistent with Falush et al. (21) suggest that genetic variation due to frequent recombination between strains is more prevalent in the non-clonal structure of the H. pylori population.
Host, nutritional and environmental factors and different H. pylori strains are effective in the results of studying H. pylori infection. Genetic studies have shown that this bacterium is very diverse, due to geographical and racial structures; for example, H. pylori strains isolated from South Asia (Korea and Singapore) have been different from Europe. The biogeographic relationship in H. pylori is most likely due to intra-family transmission (16). Generally, the distribution and prevalence of gastric cancer are dependent on H. pylori groups defined by MLST-based population analysis (31). The genetic analysis of the bacteria has shown that bacterial clones are responsible for the spread of the disease and the increased incidence of infection. Separate clones are identified frequently by a unique combination of alleles and virulence-related genes. These observations have important implications for understanding infectious diseases and measuring public health in order to reduce its adverse effects on the community.
Infected people with csga East Asian type strains have a higher risk of gastric ulcer or gastric cancer than those infected with other strains (32). The prevalence of gastric cancer is higher in regions dominated by the hpEastAsia strain, especially hspEAsia. On the other hand, the incidence of gastric cancer is very low in Africa, where most strains are hpNEAfrica, hpAfrica1 or hpAfrica2, and also in South Asia, where dominant strain is hpAsia2 (24). The strains of hpeurope and hpAfrica 1 are present together in low-risk regions. The individuals infected with the hp Europe strains of H. pylori have more histopathological degrees than those infected with hp africa1 strains. The author concluded that the differences in the bacterial population could be used to predict the risk of gastric cancer. The phylogeographic origins identified by MLST can be used to predict the risk of gastric cancer. These studies cannot determine the relationship between phylogenetic origin and clinical outcomes.
A study in Columbia found no clear link between cluster and disease. Therefore, the MLST tree topology cannot be applied as a marker in evaluating the risk for gastric cancer and digestive ulcers for hp Europe strains in Columbia. The cag A type is superior to the phylogeographic origin in predicting gastric cancer risk for hp Europe strains. Hence, the state of the virulence factors may be related to the phylogenetic tree obtained by MLST (31). The phylogeographic topology and sub-populations are significantly correlated with clinical outcomes. The difference in phylogeographic topology leads to the status of caga and vaca in the sub-populations. Therefore, the phylogeographic origins can play a role as a leading factor in predicting virulence factors but not in the outcomes of the disease.
The MLST analysis based on seven structural genes suggests that most strains isolated from Kermanshah are integrated with the population of hp Europe. The strains of hp Europe in Kermanshah are mainly induced by Spanish immigration. A typing population of H. pylori is a useful tool for mapping human migration patterns. Latifi-Navid et al. reported findings similar to our study by evaluating the of H. pylori isolated from different geographical regions of Iran (16). In a study conducted by Latifi-Navid et al. in 2010, isolated strains were close to strains isolated from Europe-Western Asia and inside the hpEurope population, which confirms the findings of the present study. The hpEurope is composed of the integration of two populations of ancestral Europe1 (AE1) in northern Europe and ancestral Europe2 (AE2) in southern Europe; their ancestral proportions are variables based on geographic location. Iranian strains showed a relatively similar distribution of the two ancestors (16).
In a study of Linz et al. (33) in 2007 in India, many strains belonged to hpAsia2. In a study by Devi et al. (34) in India in 2007, many strains belonged to hpEurope, which shows that H. pylori are more heterogeneous in the Indian population, reflecting more than two ancestors and recent migrations. It should be noted that the strains of hpAsia2 isolated from Malaysian and Ladakh Indians are divided into two sub-populations. The hspLadakh and hspIndia reflect regional differences in India. Malaysian Indians were mainly from southern India. In the present study, the sequence types were evaluated for 23 isolates, and the sequence types were new and not previously registered in the databases, indicating a high level of recombination between the isolates of H. pylori. Each isolate has different sequence types, and two pairs of isolates have common sequence types.
Liao et al. (29) in 2009 in China reported similar results to the present study. Of the 40 isolated strains, 38 new sequence types are present, and two pairs of isolates have the same sequence type. A study by Secka et al. (35) in Gombian showed that four pairs of sequence types were the same. Given that a pair of sequence types in the present study, which has the same sequence sequences, were isolated in different days of the patients and processed in the lab, thus most likely they reflect the transmission of completely related strains by unrelated members of the community. The same explanation is suggested for a pair of sequences types in the present study, which was isolated from the patients on the same day because the endoscopic devices were carefully disinfected and cleaned. Although none of these pair common strains have been separated from those with a common surname, further information is needed because they have lived in a wide common family or region and village that are affected by other factors such as hospitality to strangers and the frequent transmission of H. pylori in the developing world.
In the present study, according to the plot drawn by START v2, most of the allele numbers of H. pylori strains are unique, and the isolates of 5Ke and 26Ke are most similar in the allele numbers. In the study by Liao et al. in 2009 in China, and according to the plot drawn by START v2 software, the isolates of 42, 41 as well as the isolates of 18 and 25 have the most similarity in allele numbers (29).
5.1. Conclusions
The sequence analysis of the seven central genes (kb3.5) from each strain showed new sequence types in the present study previously not recorded in the MLST database. The sequence types of 12 isolates from Kermanshah, which are already registered on the site, are different. Genetic variation is high among H. pylori strains. The mutability pattern as a result of the inter- and intra-species mutation and recombination is abundant that may lead to adaptation in the host. The genetically identical strains are in a group and create a clonal line. The non-clonal population of H. pylori strains in this study is due to high recombination among strains that were placed within the population of hpEurope through analyzing by neighbor-joining method. The genetic characteristics of H. pylori in Iran are strongly influenced by the genetic exchanges with neighboring countries. Due to the uniqueness of the strains and their diverse sequences, the strains were placed in separate clones, and none of them could have the role of type strain in Kermanshah.
References
-
1.
Park SA, Ko A, Lee NG. Stimulation of growth of the human gastric pathogen Helicobacter pylori by atmospheric level of oxygen under high carbon dioxide tension. BMC Microbiol. 2011;11:96. [PubMed ID: 21569333]. [PubMed Central ID: PMC3110553]. https://doi.org/10.1186/1471-2180-11-96.
-
2.
Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347(15):1175-86. [PubMed ID: 12374879]. https://doi.org/10.1056/NEJMra020542.
-
3.
Bergman M, Del Prete G, van Kooyk Y, Appelmelk B. Helicobacter pylori phase variation, immune modulation and gastric autoimmunity. Nat Rev Microbiol. 2006;4(2):151-9. [PubMed ID: 16415930]. https://doi.org/10.1038/nrmicro1344.
-
4.
Isaacson PG, Du MQ. Gastrointestinal lymphoma: Where morphology meets molecular biology. J Pathol. 2005;205(2):255-74. [PubMed ID: 15643667]. https://doi.org/10.1002/path.1703.
-
5.
Kansau I, Raymond J, Bingen E, Courcoux P, Kalach N, Bergeret M, et al. Genotyping of Helicobacter pylori isolates by sequencing of PCR products and comparison with the RAPD technique. Res Microbiol. 1996;147(8):661-9. [PubMed ID: 9157493]. https://doi.org/10.1016/0923-2508(96)84023-X.
-
6.
Amirhooshang A, Ramin A, Ehsan A, Mansour R, Shahram B. High frequency of Helicobacter pylori DNA in drinking water in Kermanshah, Iran, during June-November 2012. J Water Health. 2014;12(3):504-12. [PubMed ID: 25252354]. https://doi.org/10.2166/wh.2013.150.
-
7.
Weyermann M, Rothenbacher D, Brenner H. Acquisition of Helicobacter pylori infection in early childhood: Independent contributions of infected mothers, fathers, and siblings. Am J Gastroenterol. 2009;104(1):182-9. [PubMed ID: 19098867]. https://doi.org/10.1038/ajg.2008.61.
-
8.
Vakil N, Zullo A, Ricci C, Hassan C, Vaira D. Duplicate breath testing to confirm eradication of Helicobacter pylori: Incremental benefit and cost in 419 patients. Aliment Pharmacol Ther. 2008;28(11-12):1304-8. [PubMed ID: 18774949]. https://doi.org/10.1111/j.1365-2036.2008.03846.x.
-
9.
Tay CY, Mitchell H, Dong Q, Goh KL, Dawes IW, Lan R. Population structure of Helicobacter pylori among ethnic groups in Malaysia: recent acquisition of the bacterium by the Malay population. BMC Microbiol. 2009;9:126. [PubMed ID: 19538757]. [PubMed Central ID: PMC2708179]. https://doi.org/10.1186/1471-2180-9-126.
-
10.
Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19(3):449-90. [PubMed ID: 16847081]. [PubMed Central ID: PMC1539101]. https://doi.org/10.1128/CMR.00054-05.
-
11.
Achtman M, Azuma T, Berg DE, Ito Y, Morelli G, Pan ZJ, et al. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32(3):459-70. [PubMed ID: 10320570]. https://doi.org/10.1046/j.1365-2958.1999.01382.x.
-
12.
Yamaoka Y. Helicobacter pylori typing as a tool for tracking human migration. Clin Microbiol Infect. 2009;15(9):829-34. [PubMed ID: 19702588]. [PubMed Central ID: PMC3128245]. https://doi.org/10.1111/j.1469-0691.2009.02967.x.
-
13.
Li WH, Sadler LA. Low nucleotide diversity in man. Genetics. 1991;129(2):513-23. [PubMed ID: 1743489]. [PubMed Central ID: PMC1204640].
-
14.
Suerbaum S, Smith JM, Bapumia K, Morelli G, Smith NH, Kunstmann E, et al. Free recombination within Helicobacter pylori. Proc Natl Acad Sci U S A. 1998;95(21):12619-24. [PubMed ID: 9770535]. [PubMed Central ID: PMC22880]. https://doi.org/10.1073/pnas.95.21.12619.
-
15.
Kersulyte D, Chalkauskas H, Berg DE. Emergence of recombinant strains of Helicobacter pylori during human infection. Mol Microbiol. 1999;31(1):31-43. [PubMed ID: 9987107]. https://doi.org/10.1046/j.1365-2958.1999.01140.x.
-
16.
Latifi-Navid S, Ghorashi SA, Siavoshi F, Linz B, Massarrat S, Khegay T, et al. Ethnic and geographic differentiation of Helicobacter pylori within Iran. PLoS One. 2010;5(3). e9645. [PubMed ID: 20339588]. [PubMed Central ID: PMC2842290]. https://doi.org/10.1371/journal.pone.0009645.
-
17.
Nouraie M, Latifi-Navid S, Rezvan H, Radmard AR, Maghsudlu M, Zaer-Rezaii H, et al. Childhood hygienic practice and family education status determine the prevalence of Helicobacter pylori infection in Iran. Helicobacter. 2009;14(1):40-6. [PubMed ID: 19191895]. https://doi.org/10.1111/j.1523-5378.2009.00657.x.
-
18.
Malaty HM. Epidemiology of Helicobacter pylori infection. Best Pract Res Clin Gastroenterol. 2007;21(2):205-14. [PubMed ID: 17382273]. https://doi.org/10.1016/j.bpg.2006.10.005.
-
19.
Smith JM, Smith NH, O'Rourke M, Spratt BG. How clonal are bacteria? Proc Natl Acad Sci U S A. 1993;90(10):4384-8. [PubMed ID: 8506277]. [PubMed Central ID: PMC46515]. https://doi.org/10.1073/pnas.90.10.4384.
-
20.
Josenhans C, Beier D, Linz B, Meyer TF, Suerbaum S. Pathogenomics of helicobacter. Int J Med Microbiol. 2007;297(7-8):589-600. [PubMed ID: 17416549]. https://doi.org/10.1016/j.ijmm.2007.02.006.
-
21.
Falush D, Wirth T, Linz B, Pritchard JK, Stephens M, Kidd M, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299(5612):1582-5. [PubMed ID: 12624269]. https://doi.org/10.1126/science.1080857.
-
22.
Cooper JE, Feil EJ. Multilocus sequence typing--what is resolved? Trends Microbiol. 2004;12(8):373-7. [PubMed ID: 15276613]. https://doi.org/10.1016/j.tim.2004.06.003.
-
23.
Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: A portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95(6):3140-5. [PubMed ID: 9501229]. [PubMed Central ID: PMC19708]. https://doi.org/10.1073/pnas.95.6.3140.
-
24.
Suzuki R, Shiota S, Yamaoka Y. Molecular epidemiology, population genetics, and pathogenic role of Helicobacter pylori. Infect Genet Evol. 2012;12(2):203-13. [PubMed ID: 22197766]. [PubMed Central ID: PMC3294018]. https://doi.org/10.1016/j.meegid.2011.12.002.
-
25.
Govorun VM, Lokhov PG, Moshkovskii SA, Momynaliev KT, Selesnyova OV, Kudryavtseva LV, et al. Comparative analysis of different typing methods for Helicobacter pylori clinical isolates. Biochemistry (Mosc). 2004;69(5):536-41. [PubMed ID: 15193128]. https://doi.org/10.1023/B:BIRY.0000029852.55796.af.
-
26.
Gill P, Alvandi AH, Abdul-Tehrani H, Sadeghizadeh M. Colorimetric detection of Helicobacter pylori DNA using isothermal helicase-dependent amplification and gold nanoparticle probes. Diagn Microbiol Infect Dis. 2008;62(2):119-24. [PubMed ID: 18599249]. https://doi.org/10.1016/j.diagmicrobio.2008.05.003.
-
27.
Singh A, Goering RV, Simjee S, Foley SL, Zervos MJ. Application of molecular techniques to the study of hospital infection. Clin Microbiol Rev. 2006;19(3):512-30. [PubMed ID: 16847083]. [PubMed Central ID: PMC1539107]. https://doi.org/10.1128/CMR.00025-05.
-
28.
Enright MC, Spratt BG. A multilocus sequence typing scheme for Streptococcus pneumoniae: Identification of clones associated with serious invasive disease. Microbiology. 1998;144 ( Pt 11):3049-60. [PubMed ID: 9846740]. https://doi.org/10.1099/00221287-144-11-3049.
-
29.
Liao YL, Guo G, Mao XH, Xie QH, Zhang WJ, Liu XF, et al. Core genome haplotype diversity and vacA allelic heterogeneity of Chinese Helicobacter pylori strains. Curr Microbiol. 2009;59(2):123-9. [PubMed ID: 19452214]. https://doi.org/10.1007/s00284-009-9407-x.
-
30.
Owen RJ, Xerry J. Tracing clonality of Helicobacter pylori infecting family members from analysis of DNA sequences of three housekeeping genes (ureI, atpA and ahpC), deduced amino acid sequences, and pathogenicity-associated markers (cagA and vacA). J Med Microbiol. 2003;52(Pt 6):515-24. [PubMed ID: 12748272]. https://doi.org/10.1099/jmm.0.04988-0.
-
31.
Shiota S, Suzuki R, Matsuo Y, Miftahussurur M, Tran TT, Binh TT, et al. Helicobacter pylori from gastric cancer and duodenal ulcer show same phylogeographic origin in the Andean region in Colombia. PLoS One. 2014;9(8). e105392. [PubMed ID: 25121764]. [PubMed Central ID: PMC4133377]. https://doi.org/10.1371/journal.pone.0105392.
-
32.
Matsunari O, Shiota S, Suzuki R, Watada M, Kinjo N, Murakami K, et al. Association between Helicobacter pylori virulence factors and gastroduodenal diseases in Okinawa, Japan. J Clin Microbiol. 2012;50(3):876-83. [PubMed ID: 22189111]. [PubMed Central ID: PMC3295155]. https://doi.org/10.1128/JCM.05562-11.
-
33.
Linz B, Balloux F, Moodley Y, Manica A, Liu H, Roumagnac P, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445(7130):915-8. [PubMed ID: 17287725]. [PubMed Central ID: PMC1847463]. https://doi.org/10.1038/nature05562.
-
34.
Devi SM, Ahmed I, Francalacci P, Hussain MA, Akhter Y, Alvi A, et al. Ancestral European roots of Helicobacter pylori in India. BMC Genomics. 2007;8:184. [PubMed ID: 17584914]. [PubMed Central ID: PMC1925095]. https://doi.org/10.1186/1471-2164-8-184.
-
35.
Secka O, Moodley Y, Antonio M, Berg DE, Tapgun M, Walton R, et al. Population genetic analyses of Helicobacter pylori isolates from Gambian adults and children. PLoS One. 2014;9(10). e109466. [PubMed ID: 25310300]. [PubMed Central ID: PMC4195673]. https://doi.org/10.1371/journal.pone.0109466.