Abstract
Background:
Pathogenic Escherichia coli is responsible for serious diseases; i.e.: Peritonitis, colitis, and urinary tract infections (UTIs) and even cancer, resulting in human morbidity and mortality. Environmental strains are increasingly spreading through food and dairy products, contributing to the pathogenetic burden of E. coli infections.Objectives:
This study was performed to compare phylogeny, virulence factors, pathogenicity islands (PAIs), and pathotypes in- between clinical and environmental E. coli isolates.Methods:
A total of 105 clinical (72) and environmental (33) E. coli isolates were collected. All isolates were subjected to phylogenetic typing using a new quadruplex polymerase chain reaction (PCR). Wide array of virulence genes (VGs) and PAI markers were assessed for both subtypes, as well as, the distribution of different pathotypes among the phylogenetic groups.Results:
Seven phylogenetic groups were detected; clinical isolates were more prevalent in phylogenetic groups B2 (22.2%) and D (23.6%), whereas environmental isolates were in groups A (24.2%) and B1 (60.6%). Majority of VGs were higher in clinical E. coli isolates. Environmental isolates showed higher percentage of some other VGs including; stx2 and hlyA. PAI markers were widespread among both categories, showing high extra-intestinal pathogenic E. coli (ExPEC) PAIs combination in environmental isolates. Enteropathogenic E. coli (EPEC) was the most widespread pathotype in clinical isolates versus enterohemorrhagic E. coli (EHEC) in environmental ones.Conclusions:
Escherichia coli pathogenicity armoury was not only confined to clinical isolates, but to environmental ones as well. Therefore, environmental E. coli isolates can serve as reservoirs for transmission of E. coli pathogenicity.Keywords
Escherichia coli Pathogenicity Islands Virulence Factors Polymerase Chain Reaction Serotyping
1. Background
Escherichia coli is a double-edged bacteria, ranging from being a fundamental and beneficial micro-biota in the digestive tract to an opportunistic pathogen causing intestinal and extra intestinal diseases (1, 2). Escherichia coli is the main etiological agent of urinary tract infections (UTIs) in women than in men and during childhood (3). Escherichia coli is also a major source for contamination of ground beef, cheese and other food kinds (4). The burden of E. coli pathogenicity became more complicated after the global emergence of antibiotic resistance and failure in treatment of hospital-acquired infections caused by the micro-organism (5). Diarrheagenic E. coli (DEC) and extra intestinal pathogenic E. coli (ExPEC) are the two major categories interspersed with eight E. coli pathovars (6). Six pathovars; enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC), enteroinvasive E. coli (EIEC), enteroaggregative E. coli (EAEC), and diffusely adherent E. coli (DAEC) are DEC; whereas, uropathogenic E. coli (UPEC) and neonatal meningitis E. coli (NMEC) are ExPEC (7).
Clermont et al. described a phylogenetic approach to classify E. coli into four groups A, B1, B2, and D through detection of three genes chuA, yjaA, and TspE4.C2 using triplex polymerase chain reaction (PCR) (8). A more recent categorization of E. coli strains, including an extra gene, arpA, was developed, subdividing them into eight phylogroups A, B1, B2, C, D, E, F, and clade I based on quadruplex PCR (9).
Pathogenic E. coli shares a multitude of virulence factors, enabling them to cause infections within the host. Virulence traits such as adhesions; encoded by fimH (Type 1 fimbriae), papC and papG (P fimbriae), sfaS (S fimbriae), and afa (a fimbrial adhesion) are the most critical types precisely among UPEC, enabling them to adhere to uro epithelial cells. Toxins in E. coli include heat-stable (ST a-b), heat labile (LT I-II), Shiga toxins (Stx 1-2), and hemolysin (3). Virulence genes (VGs) are often located on transmissible genetic elements called pathogenicity associated islands (PAIs). Pathogenicity island may range from 10 to 200 kb and encode genes, which are related to one or more virulence factors (10). The presence of a single or few VGs rarely endows a strain with a pathogenic status, unless that strain has acquired suitable combination of VGs. Unfortunately, acquisition of appropriate VGs combination, particularly PAIs by commensal E. coli, may qualify them to be pathogenic (11).
2. Objectives
Our study is conducted to determine the new phylogroups of E. coli isolates, the distribution of different pathotypes in each phylogroup and the pathogenic potential of environmental isolates in comparison with clinical ones through detection of a wide array of VGs and PAI markers. To our knowledge, this is the first study conducted in Egypt to investigate phylogenecity and pathogenicity of E. coli isolates from different environmental sources, based on their VGs repertoire.
3. Methods
3.1. Ethics Statement
The experimental protocol conducted in this study complies with the ethical guidelines adopted by "The Research Ethics Committee, Faculty of Pharmacy, Mansoura University” which is in accordance with the Code of Ethics of World Medical Association (Declaration of Helsinki involving use and handling of human subjects) (Permit Number: 2015-58).
3.2. Bacterial Isolates
A total of 285 clinical specimens were collected from urine (n = 107 isolates), rectum (n = 78 isolates), and surgical wound (n = 100 isolates). The specimens were obtained from local hospitals in Mansoura, Dakahlia, Egypt, namely; the Urology and Nephrology Center (UNC, n = 75 isolates), Mansoura University Hospitals (MUH, n = 70 isolates), Burns and Cosmetics Center (BCC, n = 50 isolates), Mansoura International Hospital (MIH, n = 34 isolates), Pediatric University Hospital (PUH, n = 42 isolates), Mansoura Emergency Hospital (MEH, n = 11 isolates), and Gastroenterology Center (GEC, n = 3 isolates). In addition, 165 environmental samples were isolated from stool samples of healthy volunteers (Microbiology and Immunology Lab., Faculty of Pharmacy, Mansoura University), meat products from different butchers’ shops and public supermarkets, and various sources of water and soil from different regions in Mansoura. All isolates were identified biochemically (12).
3.3. Molecular Method
3.3.1. Virulence Genotyping
All isolates were screened for 22 virulence-encoding genes (fimH, afa/draB, papC, papG, sfaS, kapsMTII, traT, iss, fyuA, iutA, bssS, astA, estA2, stp, eltB, ltA, eaeA, stx1, stx2, cnf1, cnf2 and hlyA), using primers (Integrated DNA Technologies, USA) listed in (Table 1). Polymerase chain reactions were conducted with programmed cycling conditions starting with; initial denaturation at 95°C for 5 minutes; 40 cycles, each consisting of denaturation at 95°C for 30 seconds and annealing according to temperature specified in (Table 1) for 30 seconds; extension at 72°C for 1 minute, and the reaction was ended by final extension at 72°C for five minutes.
List of Oligonucleotide Primers Used in the Study
| Genes and Primers | Nucleotide Sequence (5’ - 3’) | Amplicon Size, bp | Annealing Temp., °C | Reference | |
|---|---|---|---|---|---|
| Virulence Genes | |||||
| Adhesion | |||||
| fimH | 640 | 60 | (13) | ||
| F | TACTGCTGATGGGCTGGTC | ||||
| R | GCCGGAGAGGTAATACCCC | ||||
| afa/drABC | 380 | 68 | (14) | ||
| F | ACCCGACGCCGTTTTACATCAACCTG | ||||
| R | CCCTTCCCGCCACCTTTCAGCA | ||||
| papC | 319 | 52 | (14) | ||
| F | TGGATTGTCAGCCTCAAGGTCTA | ||||
| R | CACTGACGCCGAAAGACGTA | ||||
| papG | 1070 | 55 | (15) | ||
| F | CTGTAATTACGGAAGTATTTCTG | ||||
| R | ACTATCCGGCTCCGGATAAACCAT | ||||
| sfaS | 240 | 55 | (15) | ||
| F | GTGGATACGACGATTACTGTG | ||||
| R | CCGCCAGCATTCCCTGTATTC | ||||
| Toxin | |||||
| astA | 390 | 50 | (16) | ||
| F | GCCATCAACACAGTATATCC | ||||
| R | GAGTGACGGCTTTGTAGTCC | ||||
| estA2 | 300 | 53 | (17) | ||
| F | CGCTCAGGATGCTAAACCA | ||||
| R | AATTCACAGCAGTAATTGCT | ||||
| stp | 300 | 53 | (18) | ||
| F | TCTTTCCCCTCTTTTAGTCAG | ||||
| R | ACAGGCAGGATTACAACAAG | ||||
| eltB | 400 | 47 | (19) | ||
| F | ACGGCGTTACTATCCTCTC | ||||
| R | TGGTCTCGGTCAGATATGTG | ||||
| ltA | 200 | 49 | (20) | ||
| F | GGCGACAGATTATACCGTGC | ||||
| R | CGGTCTCTATATTCCCTGTT | ||||
| stx1 | 500 | 55.5 | (16) | ||
| F | ATAAATCGCCATTCGTTGACTAC | ||||
| R | AGAACGCCCACTGAGATCATC | ||||
| stx2 | 213 | 55.5 | (16) | ||
| F | GGCACTGTCTGAAACTGCTCC | ||||
| R | TCGCCAGTTATCTGACATTCTG | ||||
| eaeA | 550 | 55.5 | (16) | ||
| F | GACCCGGCACAAGCATAAGC | ||||
| R | CCACCTGCAGCAACAAGAGG | ||||
| cnf1 | 498 | 55 | (15) | ||
| F | AGGATGGAGTTTCCTATGCAGGAG | ||||
| R | CATTCAGAGTCCTGCCCTCATTATT | ||||
| cnf2 | 543 | 55 | (21) | ||
| F | AATCTAATTAAAGAGAAC | ||||
| R | CATGCTTTGTATATCTA | ||||
| hlyA | 1177 | 55 | (15) | ||
| F | AACAAGGATAAGCACTGTTCTGGC | ||||
| R | ACCATATAAGCGGTCATTCCCGTC | ||||
| Serum resistance | |||||
| traT | 290 | 55 | (15) | ||
| F | GGTGTGGTGCGATGAGCACAG | ||||
| R | CACGGTTCAGCCATCCCTGAG | ||||
| iss | 260 | 50 | (13) | ||
| F | GGCAATGCTTATTACAGGATGTGC | ||||
| R | GAGCAATATACCCGGGCTTCC | ||||
| kapsMTII | 269 | 60 | (14) | ||
| F | GCGCATTTGCTGATACTGTTG | ||||
| R | CATCCAGACGATAAGCATGAGC | ||||
| Iron chelation system | |||||
| iutA | |||||
| F | ATCAGAGGGACCAGCACGC | 253 | 60 | (14) | |
| R | TTCAGAGTCAGTTTCATGCCGT | ||||
| fyuA | |||||
| F | ATACCACCGCTGAAACGCTG | 277 | 60 | (14) | |
| R | CGCAGTAGGCACGATGTTGTA | ||||
| Biofilm encoding gene | |||||
| bssS | |||||
| F | GATTCAATTTTGGCGATTCCTGC | 225 | 60 | (13) | |
| R | TAATGAAGTCATTCAGACTCATCC | ||||
| Pathogenicity Islands (ExPEC PAIs) | |||||
| PAI I536 | 1800 | 55 | (22) | ||
| F | TAATGCCGGAGATTCATTGTC | ||||
| R | AGGATTTGTCTCAGGGCTTT | ||||
| PAI II536 | 1000 | 55 | (22) | ||
| F | CATGTCCAAAGCTCGAGCC | ||||
| R | CTACGTCAGGCTGGCTTTG | ||||
| PAI III536 | 200 | 55 | (22) | ||
| F | CGGGCATGCATCAATTATCTTTG | ||||
| R | TGTGTAGATGCAGTCACTCCG | ||||
| PAI IV536 | 300 | 55 | (22) | ||
| F | AAGGATTCGCTGTTACCGGAC | ||||
| R | TCGTCGGGCAGCGTTTCTTCT | ||||
| PAI ICFT073 | 930 | 56 | (22) | ||
| F | GGACATCCTGTTACAGCGCGCA | ||||
| R | TCGCCACCAATCACAGCGAAC | ||||
| PAI IICFT073 | 400 | 56 | (22) | ||
| F | ATGGATGTTGTATCGCGC | ||||
| R | ACGAGCATGTGGATCTGC | ||||
| PAI IJ96 | 400 | 53 | (22) | ||
| F | TCGTGCTCAGGTCCGGAATTT | ||||
| R | TGGCATCCCACATTATCG | ||||
| PAI IIJ96 | 2300 | 53 | (22) | ||
| F | GGATCCATGAAAACATGGTTAATGGG | ||||
| R | GATATTTTTGTTGCCATTGGTTACC | ||||
| Pathogenicity Islands (DEC PAIs) | |||||
| HPI (irp2) | 287 | 61 | (23) | ||
| F | AAGGATTCGCTGTTACCGGAC | ||||
| R | TCGTCGGGCAGCGTTTCTTCT | ||||
| Tia (tia) | 507 | 58 | (24) | ||
| F | CCCTTCTGCATCCTTGTAAGACA | ||||
| R | TATAAGGGCGGTGATAAAAACG | ||||
| O-islands (efa/lifA) | 521 | 58 | (23) | ||
| F | GAACAAAGAACATTTTCACCAGTTC | ||||
| R | CTTTCAGGTGGGGAACCCG | ||||
| She (pic) | 606 | 57 | (23) | ||
| F | ATTCTTCTGGCTGGCATTCC | ||||
| R | CGGGATTAGAGACTATTGTTGC | ||||
| EspC (espC) | 453 | 54 | (23) | ||
| F | GCTCAACTAAATATTGATAATGTATG | ||||
| R | CCCAGCCCCAACCCTGAAAC | ||||
| Primers Used for Phylogenetic Quadruplex PCR | |||||
| chuA | |||||
| chuA.1b | F | ATGGTACCGGACGAACCAAC | 288 | 59 | (9) |
| chuA.2b | R | TGCCGCCAGTACCAAAGACA | 288 | 59 | (8) |
| yjaA | 211 | 59 | (9) | ||
| yjaA.1b | F | CAAACGTGAAGTGTCAGGAG | |||
| yjaA.2b | R | AATGCGTTCCTCAACCTGTG | |||
| TspE4.C2 | 152 | 59 | (9) | ||
| TspE4C2.1b | F | CACTATTCGTAAGGTCATCC | |||
| TspE4C2.2b | R | AGTTTATCGCTGCGGGTCGC | |||
| arpA | |||||
| AceK.f | F | AACGCTATTCGCCAGCTTGC | 400 | 59 | (9) |
| ArpA1.r | R | TCTCCCCATACCGTACGCTA | 400 | 59 | (25) |
| Primers Used for Duplex PCR | |||||
| Group E | |||||
| arpA | 301 | 57 | (26) | ||
| ArpAgpE.f | F | GATTCCATCTTGTCAAAATATGCC | |||
| ArpAgpE.r | R | GAAAAGAAAAAGAATTCCCAAGAG | |||
| trpA | 489 | 57 | (27) | ||
| trpBA.f | F | CGGCGATAAAGACATCTTCAC | |||
| trpBA.r | R | GCAACGCGGCCTGGCGGAAG | |||
| Group C | |||||
| trpA | 219 | 59 | (26) | ||
| trpAgpC.1 | F | AGTTTTATGCCCAGTGCGAG | |||
| trpAgpC.2 | R | TCTGCGCCGGTCACGCCC | |||
| trpA | 489 | 59 | (27) | ||
| trpBA.f | F | CGGCGATAAAGACATCTTCAC | |||
| trpBA.r | R | GCAACGCGGCCTGGCGGAAG | |||
3.3.2. Assay of Pathogenicity Island Markers
All isolates were assessed for presence of PAIs. Pathogenicity islands belonging to ExPEC were detected through duplex PCR. Diarrheagenic pathogenicity islands were screened by uniplex PCR using primers listed in (Table 1), according to the PCR protocol prescribed above.
3.3.3. Escherichia coli Phylotyping
All isolates were classified phylogenetically into eight groups; A, B1, B2, D, F, clade I (by quadruplex PCR), C, and E (by duplex PCR assay) using the designated primers listed in (Table 1).
3.4. Escherichia coli Serotyping
The isolates were serologically identified using rapid diagnostic E. coli antisera sets (DENKA SEIKEN Co., Japan), as described previously (28).
3.5. Statistical Analysis
GraphPad Prism software (version 5.01) was used for statistical analysis of the data, applying Fisher’s exact and Chi-square tests. The level of significance was set at P value < 0.05.
4. Results
4.1. Identification of Isolates
Total; 105 isolates were identified as E. coli of which, 72 isolates were from “clinical” versus 33 isolates from “environmental” sources. Thirty “clinical” isolates were from urine samples, 21 from wound and 21 from rectal swabbing. For “environmental” isolates, 15 samples were from luncheon, milk, and cheese, 13 isolates from meat, ground beef and beef burger plus five isolates from stool of healthy subjects.
4.2. Prevalence of Virulence and Toxin Genes
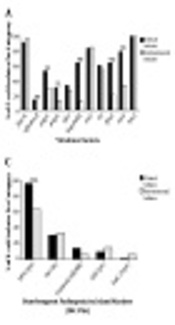
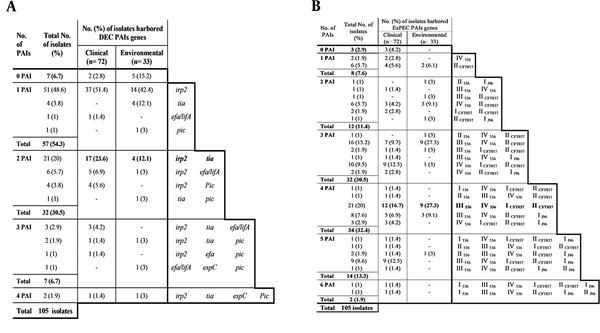
Detection of VGs and toxin genes (Figure 1A and B) revealed that, bssS, encoding biofilm, was harbored by all isolates. In contrast, cnf2 was not amplified in any isolate. Other genes were amplified by different percentages. Toxin and VGs distribution among clinical and environmental isolates showed that 12 genes (afa/draB, papC, papG, kapsMTII, fyuA, iutA, stp, estA2, eltB, ltA, eaeA, and stx2) were significantly different in-between the two isolate subtypes with P value ranging from 0.0369 to 0.0001. Furthermore, the mean score of total virulence factors among clinical isolates was 10.2, while with those of environmental ones was 7.2 (Table 2).
Distribution of; A, virulence; B, toxins; C, DEC PAIs; D, ExPEC PAIs; E, phylogroups; F, pathogroups among clinical and environmental Escherichia coli isolates. DEC, diarrheagenic E. coli; PAIs, pathogenicity associated islands; ExPEC, extra-intestinal pathogenic E. coli; EPEC, enteropathogenic E. coli; EHEC, enterohaemorrhagic E. coli; ETEC, enterotoxigenic E. coli; EIEC, enteroinvasive E. coli; *, significant; **, moderately significant; ***, highly significant.
Mean Virulence and Toxin Scores for Clinical and Environmental Escherichia coli Isolates, by Phylogenetic Groups
| Phylogenetic Group | MVSa (No. of Isolates) | MTSb (No. of Isolates) | ||||
|---|---|---|---|---|---|---|
| All Isolates (N = 105) | Clinical Isolates (N = 72) | Environmental Isolates (N = 33) | All Isolates (N = 105) | Clinical Isolates (N = 72) | Environmental Isolates (N = 33) | |
| Total | 6.1 (105) | 6.7 (72) | 4.7 (33) | 3.2 (105) | 3.5 (72) | 2.5 (33) |
| Group A | 5.5 (18) | 6.2 (10) | 4.6 (8) | 2.3 (18) | 2.3 (10) | 2.3 (8) |
| Group B1 | 4.5 (30) | 5.9 (10) | 4.3 (20) | 2.7 (30) | 3.0 (10) | 2.5 (20) |
| Group B2 | 8.6 (18) | 8.5 (16) | 9 (2) | 2.8 (18) | 2.7 (16) | 3.5 (2) |
| Group D | 6.6 (19) | 6.8 (17) | 5 (2) | 4.4 (19) | 4.6 (17) | 2.5 (2) |
| Group C | 5.6 (12) | 5.6 (12) | - | 3.9 (12) | 3.9 (12) | - |
| Group E | 5 (3) | 5.0 (3) | - | 4.3 (3) | 4.3 (3) | - |
| Group F | 7 (3) | 7.5 (2) | 6 (1) | 3.7 (3) | 4 (2) | 3 (1) |
| Unknowns | 7.5 (2) | 7.5 (2) | - | 3.5 (2) | 3.5 (2) | - |
4.3. Distribution of PAIs
Seventy one clinical isolates and all environmental isolates carried PAIs. Distribution of PAIs among clinical and environmental isolates revealed that, PAI I536, PAI III536, PAI ICFT073, PAI IJ96, and the high pathogenicity island (HPI) were significantly different in-between isolates from the two sources with P value ranging from 0.0369 to 0.0001 (Figure 1C and D). The distribution of DEC PAI markers (Figure 1C) showed that HPI (irp2) was the most common among clinical (95.8%) and environmental (63.6%) isolates. In contrast, EspC (espC) was the least detected marker in both subtypes. For ExPEC PAIs, PAI IICFT073 was the most predominant marker in both clinical (90.3%) and environmental (97%) isolates. It was noticed that, PAI I536 and PAI IIJ96 were the least ExPEC PAIs, detected only in clinical isolates in percentages of 5.6% and 2.8%, respectively (Figure 1D).
4.4. Phylogenetic Analysis
Phylogenetic distribution among all isolates is presented in Figure 1E. For clinical E. coli isolates, the distribution of phylogenetic groups was as follows: D; 23.6% (17 isolates), B2; 22.2% (16 isolates), A and B1; 13.9 % (10 isolates for each). Whereas, the distribution of environmental phylogroups was: B1; 60.6% (20 isolates), A; 24.2% (8 isolates), 6.1 % (two isolates) for both B2 and D. Eighteen isolates were found to belong to the newly assigned phylogroups; C, E, and F. The distribution of E. coli isolates in B1, B2, D, and C phylogroups was statistically significant between clinical and environmental categories (P < 0.01). For environmental isolates, the distribution in B1 group was significantly higher than that in group A (P < 0.001). Hospital distribution among each phylogroup is presented in Appendix 1 in Supplementary File. It was found that, isolates from MIH were distributed within the whole groups, unlike MUH-collected isolates, which were limited to B2, D and C groups. Most isolates of groups D and C were from UNC and MUH, respectively, while, all isolates in group E were solely from MIH.
4.5. Escherichia coli Pathotypes
Serological recognition of isolates clarified that, EPEC was the most widespread pathotype (41%), with the most abundant serotype O15: H2, while EIEC represented the least common pathotype (7.6%), with all belonging to O124 serotype. Most clinical isolates (44.4%) were EPEC, whereas, 42.4% of environmental isolates were EHEC (Figure 1F).
4.6. Pathogenicity Islands Combinations
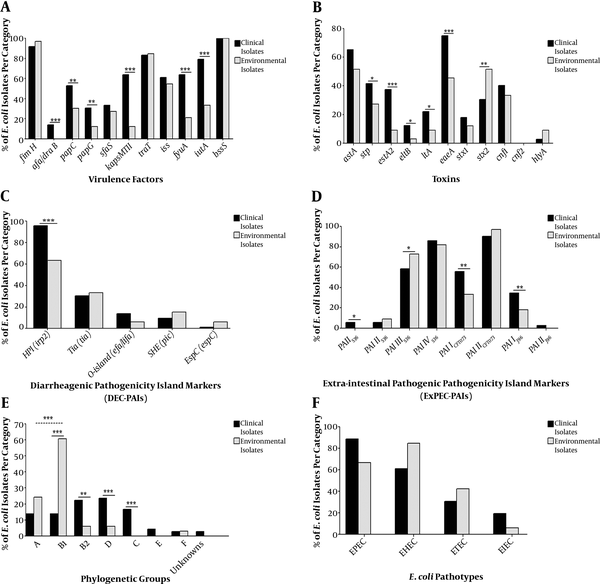
Among the environmental isolates, 27.3% (9/33 isolates) had more than one DEC PAI, compared with 44.4% (32/72 isolates) for clinical isolates (Figure 2A). In contrast, accumulation of ExPEC PAIs was strikingly high in environmental isolates (93.9%, 31/33 isolates) as compared to those in clinical ones (87.5%, 63/72 isolates, Figure 2B).
Hierarchical diagram of pathogenicity island markers among 72 clinical and 33 environmental isolates based on non-possession or possession of single/multiple combinations of pathogenicity island markers. PAIs, pathogenicity associated islands. A, diarrheagenic PAI combination; B, extra-intestinal PAI combinations.
4.7. Phylogenetic Relationship with Virulence and Toxin Genes, Pathogenicity Islands and Pathotypes
As shown in Table 2, aggregate virulence factor scores differed considerably among the phylogenetic groups. Group B2 harbored a multitude of VGs and recorded the highest mean virulence score (MVS) (the sum of all VGs detected in isolates/ the number of isolates in each category per each phylogroup) totally and within each category. Regarding the mean toxin score (MTS) (the sum of all toxin genes detected in isolates/ the number of isolates in each category per each phylogroup), D and B2 phylogroups exhibited the highest score among clinical and environmental isolates, respectively, while, groups C, E, and F exhibited intermediate scores, and group A exhibited the lowest one.
The relation between phylogenetic classification and PAIs was evaluated by means of median PAIs as shown in Figure 3A and B (number of isolates with PAIs/ total number of isolates in each category per each phylogroup). The highest median DEC PAIs (Figure 3A) was recorded for clinical isolates of phylogroup C (20/12 isolates) and phylogroup E (5/3 isolates), whereas, for environmental isolates, it was in D group (4/2 isolates). Group B2 exhibited the highest median ExPEC PAI in both clinical and environmental populations (Figure 3B), with somewhat greater proportion in environmental isolates (9/2) versus that for clinical ones (66/16). Regarding inter-relationship between phylogeny and pathotypes (Figure 3C), it was found, that EPEC isolates fall predominantly in group D (23.3%), and all of them were clinically-isolated. Twenty five percent of EHEC and 55.6% of ETEC were intensely connected to the B1 phylogroup. The least detectable pathotype, EIEC, was distributed equally among A, B1, and B2 groups (two isolates each), in addition to D and C groups (one isolate each).
5. Discussion
Most E. coli strains can colonize in the intestinal tract with no harmful effects to the host, however, some strains are pathogenic. The growth of E. coli in soil, sediment, and water has been widely documented. Escherichia coli isolates have also been found in dairy products (29). In our study, detection of virulence and toxin genes suggested the pathogenicity of environmental isolates. Although 11 genes (afa/draB, papC, papG, kapsMTII, fyuA, iutA, stp, estA2, eltB, ltA and eaeA) were found to be significantly higher in clinical isolates as compared to environmental ones (Figure 1A and B), the percentage is still high in environmental isolates suggesting potential virulence. The prevalence of VGs revealed that, cnf2 was absent in all isolates, which came in agreement with Baliere et al. (30). Conversely, bssS gene was carried by all isolates, reflecting their potential to survive and attach to surfaces (31).
Among the adhesion genes, fimH was the most prevalent in both clinical and environmental isolates. The high binding ability of fimH and its importance in colonizing different niches can result in increased pathogenicity of E. coli (32). Of genes conferring serum resistance, traT was the most predominant in both isolates subtypes. The acquisition of environmental E. coli isolates for comparable levels of fimH and traT genes to clinical isolates, may suggest their pathogenicity and ability to cause host illness (33). On the other hand, iutA gene, which encodes for a virulence factor contributing to bacterial growth where iron availability is limited (34), was considered the most prevalent siderophore in both isolates subtypes. Clinical isolates showed higher percentage for all VGs, except stx2 and hlyA, which were interestingly higher in environmental isolates. This can be interpreted as a result of poor hygiene for food handlers or environmental samples contamination by livestock manure, wastewaters, or by wildlife (35).
Since E. coli pathogenicity is linked to PAI markers, our study evaluated both DEC and ExPEC PAIs. We have found that, HPI was the most abundant marker in both categories of our isolates (Figure 1C), agreeing with Naderi et al. (23). Substantial percentage of HPI in environmental isolates emphasized it as being “fitness” rather than “pathogenicity” island. For other DEC PAIs, EspC was the least detected marker (Figure 1C), which came in accordance with Makobe et al. (36). Unlike the high prevalence of PAI IV536 marker in the majority of studies (22, 37), our study demonstrated that PAI IICFT073 was the most prevalent marker in both categories (Figure 1D). Moreover, the presence of PAI III536 was high as compared to PAI II536, agreeing with Dobrindt et al. (38). PAI I536 and PAI IIJ96 were the least common markers and were distributed only in clinical isolates (Figure 1D). This can be explained as acquisition and stabilization of PAI I536 and PAI IIJ96 on the chromosome was very low, so isolates carrying them were rare and their presence was limited only to highly virulent strains (22).
The presence of multiple PAI markers among isolates is important for estimating its virulence potential (23). We found that combination of DEC PAI markers was greater in clinical isolates since the great majority of environmental isolates (72.7% ) either does not or harbors only one DEC PAI (Figure 2A). The reverse scenario was observed for ExPEC PAIs combination, where dominant percentage was observed in environmental isolates (Figure 2B). The unusual PAIs prevalence among environmental isolates may provide them with means for infection, reaching sepsis (35). With respect to median PAIs (Figure 3A and B), there was very close convergence between clinical and environmental isolates. These results substantiate the probability that environmental isolates are not commensals, which came in accordance with Koga et al. (39).
Comparison between clinical and environmental Escherichia coli isolates via relation between phylogeny and A, median DEC PAIs; B, median ExPEC PAIs; C, pathotypes. PAIs, pathogenicity associated islands; solid shapes, indicate clinical isolates; empty shapes, indicate environmental isolates.
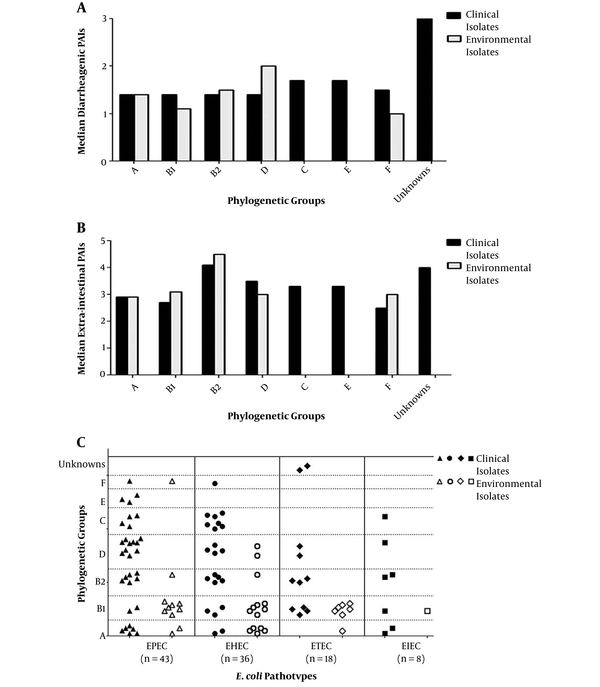
Phylogenetic analysis is a key to examine E. coli diversity and understand their characteristics in order to establish control programs and settle alterative therapeutic options (40). Our results revealed that, B2 and D groups were the predominant phylogroups in clinical isolates, whereas, A and B1 groups were the most populous among environmental ones (Figure 1E). Around 17% of our isolates were assigned as new phylogroups; C, E, and F, of which the majority were clinically isolated (17/18 isolates, 94.4%). Only 1.9% of our isolates were unassigned to any of the phylogenetic groups, agreeing with Clermont et al. (9).
The relationship of phylogeny with virulence machinery, examined in our study (Table 2), demonstrated firm tying of VGs to B2 and D groups, asserting previous findings (41, 42). First; by taking the environmental total virulence factor score (7.2) as a scale, 61 clinical isolates were considered highly virulent, harboring more than seven VGs and they were more predominant in D and B2 groups. Second; three isolates code (W6, E7 and E25), harboring the highest number of tested genes, were in B2 group. Third; more than half of tested VGs (12 gene) were found to be associated with B2 and D groups. Fourth; B2 and D phylogroups registered the highest MVS, MTS and median ExPEC PAIs, totally and within each category. In terms of MVS, B2 was the highest group followed by the F group. As for MTS, D was the highest group followed by C and E groups. Moreover, C and E phylogroups recorded the highest median DEC PAI (Fig 3A). These results proposed that C, E and F phylogenetic types were a successor for the highly pathogenic B2 and D groups.
Environmental isolates, harboring high VGs (more than seven, 33.3%), were surprisingly located in A and B1 groups. Since these isolates can be considered as reservoirs for pathogenicity, we can conclude that, not all isolates in A and B1 groups are harmless commensals. Our results came in agreement with Koga et al. (39). Our isolates were allocated to different pathotypes, reflecting their different features of pathogenesis. All pathotypes were identified except EAEC and DAEC (Figure 1F). Enteropathogenic E. coli was the most prevalent pathotype, while, EIEC was the least common one, indicating that EIEC is not locally virulent in Egypt. Since most environmental isolates were EHEC, they are considered a gateway for dysentery and hemolytic uremic syndrome (43). This matches the results of Robbins et al. who demonstrated ground beef contamination with Shiga toxin-producing E. coli (STEC) (44). Detection of ETEC in environmental isolates was surprising, as ETEC is recognized as a waterborne more than a foodborne pathogen (45). According to the phylogenetic results (Figure 3C), 23.3% of EPEC fell in D group, while, 25% of EHEC isolates were in B1 group. Finally, serological classification of E. coli affirmed that not all environmental isolates were avirulent. Nearly all pathovars were detected in our environmental isolates and furthermore can transmit ETEC and possibly EPEC and STEC, agreeing with Amezquita-Montes et al. (46). Indeed, most studies reported that isolates of phylogroups A and B1 are commensals (47, 48), however, this can be judged as, per our results, B1 group was found to be equipped with the most critical pathotypes; EHEC and ETEC.
5.1. Conclusions
The results of the current study highlight the fact that, pathogenic E. coli are not only confined to hospitals, but in food and dairies as well. The newly described phylogroups; C, E, and F were not inferior to B2 and D phylogroups in their virulence potential. Escherichia coli pathogenicity determined by panoply of virulence traits, pathogenicity markers and phylogenetic architecture, was not only limited to clinical isolates, but also to environmental ones. Our findings support the fact that, environmental isolates contribute to the local spread of E. coli pathogenicity in Egypt, yet, further investigation is required to examine how wide-spread the problem is. To conclude, it is always beneficial and safer to implement proper food-handling practices in restaurants and supermarkets to limit the spread of E. coli-induced infections.
Acknowledgements
References
-
1.
Rehman MU, Zhang H, Iqbal MK, Mehmood K, Huang S, Nabi F, et al. Antibiotic resistance, serogroups, virulence genes, and phylogenetic groups of Escherichia coli isolated from yaks with diarrhea in Qinghai Plateau, China. Gut Pathog. 2017;9:24. [PubMed ID: 28546830]. [PubMed Central ID: PMC5443361]. https://doi.org/10.1186/s13099-017-0174-0.
-
2.
Najafi A, Hasanpour M, Askary A, Aziemzadeh M, Hashemi N. Distribution of pathogenicity island markers and virulence factors in new phylogenetic groups of uropathogenic Escherichia coli isolates. Folia Microbiol (Praha). 2018;63(3):335-43. [PubMed ID: 29199378]. https://doi.org/10.1007/s12223-017-0570-3.
-
3.
Sedighi I, Nejad ASM, Amanati A, Nakhaei S, Alikhani MY. Virulence factors and antibiotic resistance in uropathogenic and commensal escherichia coli isolates. J Krishna Inst Med Sci Univ. 2016;5:50-7.
-
4.
Johnson JR, Johnston BD, Delavari P, Thuras P, Clabots C, Sadowsky MJ. Phylogenetic backgrounds and virulence-associated traits of Escherichia coli isolates from surface waters and diverse animals in Minnesota and Wisconsin. Appl Environ Microbiol. 2017;83(24). [PubMed ID: 28986372]. [PubMed Central ID: PMC5717207]. https://doi.org/10.1128/AEM.01329-17.
-
5.
Haghighatpanah M, Mozaffari Nejad AS, Mojtahedi A, Amirmozafari N, Zeighami H. Detection of extended-spectrum beta-lactamase (ESBL) and plasmid-borne blaCTX-M and blaTEM genes among clinical strains of Escherichia coli isolated from patients in the north of Iran. J Glob Antimicrob Resist. 2016;7:110-3. [PubMed ID: 27721192]. https://doi.org/10.1016/j.jgar.2016.08.005.
-
6.
Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2(2):123-40. [PubMed ID: 15040260]. https://doi.org/10.1038/nrmicro818.
-
7.
Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol. 2010;8(1):26-38. [PubMed ID: 19966814]. https://doi.org/10.1038/nrmicro2265.
-
8.
Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol. 2000;66(10):4555-8. [PubMed ID: 11010916]. [PubMed Central ID: PMC92342]. https://doi.org/10.1128/AEM.66.10.4555-4558.2000.
-
9.
Clermont O, Christenson JK, Denamur E, Gordon DM. The clermont Escherichia coli phylo-typing method revisited: Improvement of specificity and detection of new phylo-groups. Environ Microbiol Rep. 2013;5(1):58-65. [PubMed ID: 23757131]. https://doi.org/10.1111/1758-2229.12019.
-
10.
Navidinia M, Peerayeh SN, Fallah F, Bakhshi B. Phylogenetic groups and pathogenicity island markers in Escherichia coli isolated from children. Jundishapur J Microbiol. 2013;6(10). https://doi.org/10.5812/jjm.8362.
-
11.
Chapman TA, Wu XY, Barchia I, Bettelheim KA, Driesen S, Trott D, et al. Comparison of virulence gene profiles of Escherichia coli strains isolated from healthy and diarrheic swine. Appl Environ Microbiol. 2006;72(7):4782-95. [PubMed ID: 16820472]. [PubMed Central ID: PMC1489375]. https://doi.org/10.1128/AEM.02885-05.
-
12.
Mahon CR, Lehman DC, Manuselis G. Textbook of diagnostic microbiology. 3rd ed. St. Louis, Mo., USA: Saunders Elsevier; 2007.
-
13.
Gharrah MM, Mostafa El-Mahdy A, Barwa RF. Association between virulence factors and extended spectrum beta-lactamase producing Klebsiella pneumoniae compared to nonproducing isolates. Interdiscip Perspect Infect Dis. 2017;2017:7279830. [PubMed ID: 28684959]. [PubMed Central ID: PMC5480045]. https://doi.org/10.1155/2017/7279830.
-
14.
Abdelmegeed ES, Barwa RF, El Galil KHA. Comparative study on prevalence and association of some virulence factors with extended spectrum beta-lactamases and AmpC producing Escherichia coli. Africa J Microbiol Res. 2015;9(17):1165-74. https://doi.org/10.5897/AJMR2015.7463.
-
15.
Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J Infect Dis. 2000;181(1):261-72. [PubMed ID: 10608775]. https://doi.org/10.1086/315217.
-
16.
Abdel-Rhman SH, Khalifa SM, El Galil KHA, Barwa RM. Prevalence of toxins and antimicrobial resistance among E. coli Isolated from meat. Adv Microbiol. 2015;5(11):737. https://doi.org/10.4236/aim.2015.511078.
-
17.
Alwayli D, Abdelmegeed E, Ibrahim RH. Studies on enterotoxins of Escherichia coli strains isolated from various sources. Egypt J Med Microbiol. 2013;36:117-35.
-
18.
Woodward MJ, Carroll PJ, Wray C. Detection of entero- and verocyto-toxin genes in Escherichia coli from diarrhoeal disease in animals using the polymerase chain reaction. Vet Microbiol. 1992;31(2-3):251-61. [PubMed ID: 1626374]. https://doi.org/10.1016/0378-1135(92)90083-6.
-
19.
Sjoling A, Wiklund G, Savarino SJ, Cohen DI, Svennerholm AM. Comparative analyses of phenotypic and genotypic methods for detection of enterotoxigenic Escherichia coli toxins and colonization factors. J Clin Microbiol. 2007;45(10):3295-301. https://doi.org/10.1128/JCM.00471-07.
-
20.
Stacy-Phipps S, Mecca JJ, Weiss JB. Multiplex PCR assay and simple preparation method for stool specimens detect enterotoxigenic Escherichia coli DNA during course of infection. J Clin Microbiol. 1995;33(5):1054-9. [PubMed ID: 7615704]. [PubMed Central ID: PMC228103].
-
21.
Blanco M, Blanco JE, Alonso MP, Blanco J. Virulence factors and O groups of Escherichia coli isolates from patients with acute pyelonephritis, cystitis and asymptomatic bacteriuria. Eur J Epidemiol. 1996;12(2):191-8. [PubMed ID: 8817199]. https://doi.org/10.1007/BF00145506.
-
22.
Sabate M, Moreno E, Perez T, Andreu A, Prats G. Pathogenicity island markers in commensal and uropathogenic Escherichia coli isolates. Clin Microbiol Infect. 2006;12(9):880-6. [PubMed ID: 16882293]. https://doi.org/10.1111/j.1469-0691.2006.01461.x.
-
23.
Naderi G, Haghi F, Zeighami H, Hemati F, Masoumian N. Distribution of pathogenicity island (PAI) markers and phylogenetic groups in diarrheagenic and commensal Escherichia coli from young children. Gastroenterol Hepatol Bed Bench. 2016;9(4):316-24. [PubMed ID: 27895858]. [PubMed Central ID: PMC5118857].
-
24.
Kariyawasam S, Johnson TJ, Debroy C, Nolan LK. Occurrence of pathogenicity island I(APEC-O1) genes among Escherichia coli implicated in avian colibacillosis. Avian Dis. 2006;50(3):405-10. [PubMed ID: 17039841]. https://doi.org/10.1637/7462-102705R.1.
-
25.
Clermont O, Bonacorsi S, Bingen E. Characterization of an anonymous molecular marker strongly linked to Escherichia coli strains causing neonatal meningitis. J Clin Microbiol. 2004;42(4):1770-2. [PubMed ID: 15071045]. [PubMed Central ID: PMC387582]. https://doi.org/10.1128/JCM.42.4.1770-1772.2004.
-
26.
Lescat M, Clermont O, Woerther PL, Glodt J, Dion S, Skurnik D, et al. Commensal Escherichia coli strains in Guiana reveal a high genetic diversity with host-dependant population structure. Environ Microbiol Rep. 2013;5(1):49-57. [PubMed ID: 23757130]. https://doi.org/10.1111/j.1758-2229.2012.00374.x.
-
27.
Clermont O, Lescat M, O'Brien CL, Gordon DM, Tenaillon O, Denamur E. Evidence for a human-specific Escherichia coli clone. Environ Microbiol. 2008;10(4):1000-6. [PubMed ID: 18177373]. https://doi.org/10.1111/j.1462-2920.2007.01520.x.
-
28.
Kok T, Worswich D, Gowans E. Some serological techniques for microbial and viral infections. Practical Medical Microbiology. 14th ed. Edinburgh, UK: Churchill Livingstone; 1996.
-
29.
Jang J, Hur HG, Sadowsky MJ, Byappanahalli MN, Yan T, Ishii S. Environmental Escherichia coli: Ecology and public health implications-A review. J Appl Microbiol. 2017;123(3):570-81. [PubMed ID: 28383815]. https://doi.org/10.1111/jam.13468.
-
30.
Baliere C, Rince A, Delannoy S, Fach P, Gourmelon M. Molecular profiling of shiga toxin-producing Escherichia coli and enteropathogenic E. coli strains isolated from french coastal environments. Appl Environ Microbiol. 2016;82(13):3913-27. [PubMed ID: 27107119]. [PubMed Central ID: PMC4907183]. https://doi.org/10.1128/AEM.00271-16.
-
31.
Abberton CL, Bereschenko L, van der Wielen PW, Smith CJ. Survival, biofilm formation, and growth potential of environmental and enteric Escherichia coli strains in drinking water microcosms. Appl Environ Microbiol. 2016;82(17):5320-31. [PubMed ID: 27342552]. [PubMed Central ID: PMC4988207]. https://doi.org/10.1128/AEM.01569-16.
-
32.
Hojati Z, Zamanzad B, Hashemzadeh M, Molaie R, Gholipour A. The FimH gene in uropathogenic Escherichia coli strains isolated from patients with urinary tract infection. Jundishapur J Microbiol. 2015;8(2). e17520. [PubMed ID: 25825648]. [PubMed Central ID: PMC4376967]. https://doi.org/10.5812/jjm.17520.
-
33.
Schmidt H, Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin Microbiol Rev. 2004;17(1):14-56. [PubMed ID: 14726454]. [PubMed Central ID: PMC321463]. https://doi.org/10.1128/CMR.17.1.14-56.2004.
-
34.
Gao Q, Wang X, Xu H, Xu Y, Ling J, Zhang D, et al. Roles of iron acquisition systems in virulence of extraintestinal pathogenic Escherichia coli: Salmochelin and aerobactin contribute more to virulence than heme in a chicken infection model. BMC Microbiol. 2012;12(1):143. https://doi.org/10.1186/1471-2180-12-143.
-
35.
Nagarjuna D, Mittal G, Dhanda RS, Verma PK, Gaind R, Yadav M. Faecal Escherichia coli isolates show potential to cause endogenous infection in patients admitted to the ICU in a tertiary care hospital. New Microb Infect. 2015;7:57-66. https://doi.org/10.1016/j.nmni.2015.05.006.
-
36.
Makobe CK, Sang WK, Kikuvi G, Kariuki S. Molecular characterization of virulence factors in diarrhoeagenic Escherichia coli isolates from children in Nairobi, Kenya. J Infect Dev Ctries. 2012;6(8):598-604. [PubMed ID: 22910565]. https://doi.org/10.3855/jidc.2082.
-
37.
Middendorf B, Hochhut B, Leipold K, Dobrindt U, Blum-Oehler G, Hacker J. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J Bacteriol. 2004;186(10):3086-96. [PubMed ID: 15126470]. [PubMed Central ID: PMC400636]. https://doi.org/10.1128/JB.186.10.3086-3096.2004.
-
38.
Dobrindt U, Blum-Oehler G, Nagy G, Schneider G, Johann A, Gottschalk G, et al. Genetic structure and distribution of four pathogenicity islands (PAI I(536) to PAI IV(536)) of uropathogenic Escherichia coli strain 536. Infect Immun. 2002;70(11):6365-72. [PubMed ID: 12379716]. [PubMed Central ID: PMC130402]. https://doi.org/10.1128/IAI.70.11.6365-6372.2002.
-
39.
Koga VL, Tomazetto G, Cyoia PS, Neves MS, Vidotto MC, Nakazato G, et al. Molecular screening of virulence genes in extraintestinal pathogenic Escherichia coli isolated from human blood culture in Brazil. Biomed Res Int. 2014;2014:465054. [PubMed ID: 24822211]. [PubMed Central ID: PMC4009324]. https://doi.org/10.1155/2014/465054.
-
40.
Mustak HK, Gunaydin E, Kaya IB, Salar MO, Babacan O, Onat K, et al. Phylo-typing of clinical Escherichia coli isolates originating from bovine mastitis and canine pyometra and urinary tract infection by means of quadruplex PCR. Vet Q. 2015;35(4):194-9. [PubMed ID: 26133976]. https://doi.org/10.1080/01652176.2015.1068963.
-
41.
Lee JH, Subhadra B, Son YJ, Kim DH, Park HS, Kim JM, et al. Phylogenetic group distributions, virulence factors and antimicrobial resistance properties of uropathogenic Escherichia coli strains isolated from patients with urinary tract infections in South Korea. Lett Appl Microbiol. 2016;62(1):84-90. [PubMed ID: 26518617]. https://doi.org/10.1111/lam.12517.
-
42.
Vargova R, Kmetova M, Curova K, Siegfried L. Virulence genes in Escherichia coli strains isolated from urine of elderly patients. Biologia. 2017;72(3):259-66. https://doi.org/10.1515/biolog-2017-0037.
-
43.
Werber D, Krause G, Frank C, Fruth A, Flieger A, Mielke M, et al. Outbreaks of virulent diarrheagenic Escherichia coli--Are we in control? BMC Med. 2012;10:11. [PubMed ID: 22300479]. [PubMed Central ID: PMC3350439]. https://doi.org/10.1186/1741-7015-10-11.
-
44.
Robbins A, Anand M, Nicholas DC, Egan JS, Musser KA, Giguere S, et al. Ground beef recall associated with non-O157 Shiga toxin-producing Escherichia coli, United States. Emerg Infect Dis. 2014;20(1):165-7. [PubMed ID: 24377832]. [PubMed Central ID: PMC3887488]. https://doi.org/10.3201/eid2001.130915.
-
45.
Daniels NA, Neimann J, Karpati A, Parashar UD, Greene KD, Wells JG, et al. Traveler's diarrhea at sea: Three outbreaks of waterborne enterotoxigenic Escherichia coli on cruise ships. J Infect Dis. 2000;181(4):1491-5. [PubMed ID: 10762583]. https://doi.org/10.1086/315397.
-
46.
Amezquita-Montes Z, Tamborski M, Kopsombut UG, Zhang C, Arzuza OS, Gomez-Duarte OG. Genetic relatedness among Escherichia coli pathotypes isolated from food products for human consumption in Cartagena, Colombia. Foodborne Pathog Dis. 2015;12(5):454-61. [PubMed ID: 25786140]. [PubMed Central ID: PMC4516910]. https://doi.org/10.1089/fpd.2014.1881.
-
47.
Carlos C, Pires MM, Stoppe NC, Hachich EM, Sato MI, Gomes TA, et al. Escherichia coli phylogenetic group determination and its application in the identification of the major animal source of fecal contamination. BMC Microbiol. 2010;10:161. [PubMed ID: 20515490]. [PubMed Central ID: PMC2889953]. https://doi.org/10.1186/1471-2180-10-161.
-
48.
Kaper JB. Pathogenic Escherichia coli. Int J Med Microbiol. 2005;295(6-7):355-6. https://doi.org/10.1016/j.ijmm.2005.06.008.