Abstract
Background:
Some evidence indicates that there is a potential linkage between ciprofloxacin resistance and the prevalence of virulence factors in pathogenic Escherichia coli strains.Objectives:
The study was conducted to evaluate the association of eight putative adhesin-encoding genes and 12 O-serotypes among ciprofloxacin susceptible/resistant in uropathogenic E. coli (UPEC) isolates.Methods:
A total of 100 E. coli isolates collected from symptomatic patients with urinary tract infection were surveyed for antimicrobial susceptibility test and polymerase chain reaction (PCR) to find the presence of eight putative adhesin-encoding genes and 12 O-serotype and their association with ciprofloxacin susceptibility and resistance.Results:
The highest and the lowest resistance rates were obtained against ampicillin (92%) and gentamycin (19%), respectively. However, the resistance rate to ciprofloxacin was detected in 43% of E. coli isolates. PCR results revealed the frequency of eight putative adhesin markers ranged from 4 to 95%. The presence of 2 genes (fimH and sfa) was >76% among all screened isolates. Of 100 UPEC isolates, 73 were positive for one of the tested O-serotypes. The most common types of O-serotype were O2 (16%), O6 (16%), and O18 (14%). The fimH, sfa, and papC were the most prevalent pattern in the ciprofloxacin-susceptible isolates (31 vs. 17). There was no significant correlation between the presence of O-serotype, adhesin-encoding genes and susceptibility/resistance to ciprofloxacin.Conclusions:
The results revealed that UPEC isolates of different geographical regions might have various properties. It is worthwhile to elucidate the differences that might result in producing valuable evidence based on clinical guidelines for the management of urinary tract infection.Keywords
Escherichia coli Adhesin-Encoding Genes O-Serotyping Ciprofloxacin
1. Background
Urinary tract infections (UTIs) are one of the most usual among community-acquired and nosocomial infections encountered by physicians (1). These infections are rarely directly associated with the death of the patients. However, they play an accelerating role in the risk of comorbidity and healthcare-associated costs. Such infectious diseases are also responsible for increase in antibiotic prescription by physicians. Fluoroquinolones and quinolone antibiotics are broad-spectrum antibiotics that are used to treat several Gram-negative and Gram-positive bacterial infections. The quinolone antibiotics were first described in the early 1960s, have become prevalent in the treatment of urinary and urogenital Escherichia coli infections (2). Quinolone-resistant E. coli (QREC) has recently emerged in different geographical regions as an important cause of extraintestinal infections (3-5).
Uropathogenic E. coli (UPEC) harbors a range of virulence determinants that relate to its ability to colonize the urinary tract and cause disease. The most significant adhesin-encoding genes include fimH, sfa, papC, foc, afa, papGI, papGII, and papGIII and genes related to the adhesin systems. Several studies have shown that these adhesin factors have significant roles in the pathogenicity of UPEC strains because the strains are able to colonize and overcome host defense systems, thereby resulting in the disease (6, 7). The increase in the multi-drug resistant strains of E. coli in many countries has caused failure in the treatment accompanied by a huge health burden (8).
A very limited range of antibacterial agents remains due to the appearance of QREC isolates, warranting that a simple case of UTI is increasingly a competing element (9). Studies have suggested that fluoroquinolone and QREC strains show fewer virulence genes and are less capable of causing infection compared with susceptible strains (10-13). In various parts of Iran, it is important to perform research studies to document the prevalence of UPEC and determine the putative virulence genes and antibiotic resistance of UPEC isolates. The complex interactions of the virulence characteristics and the O-serotype background similar to the antibiotic resistance of E. coli resulting from their various relationships need further study.
2. Objectives
The objective of the present study is to clarify the type of association of the acquisition of virulence factors (VFs) with resistance alone and resistance depends on an O-serotype or unknown factors. The antibiotic resistance, the genotypic adhesin factors and O-serotype characterization in UPEC isolates were assessed.
3. Methods
3.1. Case Definition
The UTIs are determined when the following symptoms appear: (1) bacteria with ≥ 104 CFU/mL count in mid-stream urine, (2) the observation of ≥ 5 leukocytes per high power field, and (3) the presence of symptoms of UTI (dysuria, frequency or urgency of urination) in the host.
3.2. Experiment Setting
One hundred non-repetitive E. coli isolates were collected from symptomatic UTI patients in a teaching hospital in Zabol, southeast of Iran. Among the 100 UTI diagnosed patients, 61 (61%) were females and 39 (39%) were males, with a mean age of 35.5 ± 18.3 (mean ± SD).
3.3. Antimicrobial Susceptibility Testing
The Kirby-Bauer’s disk diffusion method was applied for antibiotic susceptibility test on Mueller Hinton agar following the CLSI guidelines (14) using antibiotic discs as follows: cefazolin (CZ), ampicillin (AM), ciprofloxacin (CP), azithromycin (AZM), cefixime (CFM), ceftazidime (CAZ), cefotaxime (CTX), and gentamycin (GM) (Padtan Teb, Iran). Standard E. coli ATCC 25922 strain was used as the control.
3.4. Detection of Adhesin Genes
The DNA of overnight cultures of UPEC isolates was extracted by boiling method (15). The uniplex-PCR was used to identify the presence of eight adhesin-encoding genes: afa, fimH, foc, papC, papG alleles (papGI, papGII and papGIII), and sfa in UPEC isolates. The predicted sizes and details of the primer sequences of the amplified products are shown in Table 1. Amplification reactions were performed in a total volume of 25 µL, including 8.5 µL ddH2O, 12.5 µL of Master Mix Red (amplicon), 2 µL of template DNA, and 1 µL of each primer (30 pmol of each of the primers) (Pishgam, Iran). PCR reactions were done in a Mastercycler gradient® pro (Eppendorf, Hamburg, Germany) using the following conditions, initial denaturation for 4 min at 94ºC followed by 35 cycles of 30 s at 94ºC, 50 s at 60ºC, and 70 s at 72ºC, with a final extension step for 7 min at 72ºC.
Primer Used In This Study
| Primer Sequence (5’-3’) | Size, bp |
|---|---|
| fimH | 400 |
| GTTGTTCTGTCGGCTCTGTC | |
| TAAATGTCGCACCATCCAG | |
| papC | 328 |
| GACGGCTGTACTGCAGGGTGTGGCG | |
| ATATCCTTTCTGCAGGGATGCAATA | |
| sfa | 100 |
| CCGTAAAGATGTCTGCGAG | |
| AGCAAGTCTG¬GCAACGAG | |
| foc | 388 |
| GGTGGAACCGCAGAAAATAC | |
| GAACTGTTGGGGAAAGAGTG | |
| afa | 750 |
| GCTGGGCAGCAAACTGATAACTCTC | |
| CATCAAGCTGTTTGTTCGTCCGCCG | |
| papGI | 461 |
| TCGTGCTCAGGTCCGGAATTT | |
| TGGCATCCCCCAACATTATCG | |
| papGII | 190 |
| GGGATGAGCGGGCCTTTGAT | |
| CGGGCCCCCAAGTAACTCG | |
| papGIII | 258 |
| GGCCTGCAATGGATTTACCTGG | |
| CCACCAAATGACCATGCCAGAC | |
| gndbis.F | - |
| ATACCGACGACGCCGATCTG | |
| rfbO1.R | 189 |
| CCAGAAATACACTTGGAGAC | |
| rfbO2a.R | 274 |
| GTGACTATTTCGTTACAAGC | |
| rfbO18.R | 360 |
| GAAGATGGCTATAATGGTTG | |
| rfbO16.R | 450 |
| GGATCATTTATGCTGGTACG | |
| rfbO6a.R | 584 |
| AAATGAGCGCCCACCATTAC | |
| rfbO7.R | 722 |
| CGAAGATCATCCACGATCCG | |
| rfbO4.R | 193 |
| AGGGGCCATTTGACCCACTC | |
| rfbO12.R | 239 |
| GTGTCAAATGCCTGTCACCG | |
| rfbO25a.R | 313 |
| GAGATCCAAAAACAGTTTGTG | |
| rfbO75.R | 419 |
| GTAATAATGCTTGCGAAACC | |
| rfbO15.R | 536 |
| TGATAATGACCAACTCGACG | |
| rfbO157.R | 672 |
| TACGACAGAGAGTGTCTGAG |
3.5. UPEC Sero-Grouping
The E. coli sero-grouping was determined by the amplification of the following target genes: O1, O2, O4, O6, O7, O12, O15, O16, O18, O25, O75, and O157 as described by Clermont et al. (16). In brief, the experiment was performed in two separate PCR runs, including six reverse primers representative of six O-reverse and one universal forward primer (Table 1) as described by Clermont et al. (16). Each 25 µL PCR mixture contained 9.5 µL ddH2O, 0.2 mM/mL of each primer (1 µL) (Pishgam, Iran), 2 µL of genomic DNA and 12.5 µL of Taq DNA Polymerase Master Mix Red (amplicon). The PCR amplification was done in the following condition: 95ºC for 4 min, 30 cycles of 95ºC for 40 s, 57ºC for 30 s, and 72ºC for 30 s, with a final extension of 72ºC for 6 min. The PCR products were visualized by 1.5% agarose gel (1× TAE buffer).
3.6. Statistical Analysis
The analyses of data were done by appropriate descriptive statistics. Statistical Package for the Social Sciences (SPSS) V 16.0 software was used for statistical analysis. Descriptive analysis of data was done by chi-square and Fisher’s exact tests at the P value of ≤ 0.05.
4. Results
4.1. Antibiotic Resistance
The antibiotic susceptibility test showed that the 92%, 74%, 71%, 65%, 55%, 53%, 43%, and 19% of UPEC isolates were resistant to ampicillin, cefazolin, cefixime, cefotaxime, ceftazidime, azithromycin, ciprofloxacin, and gentamycin, respectively. According to in vitro findings, ciprofloxacin and gentamycin were more effective antibiotics for the treatment of UPEC isolates, while ampicillin, cefazolin and cefixime had the least therapeutic effects. More than 70% of E. coli isolates were resistant to the third generation of the tested cephalosporins and 57% were susceptible to ciprofloxacin (Table 2).
Pattern of Antibiotic Susceptibility and Distribution of Adhesin Genes Among Escherichia coli Isolates
| CZ | CP | AZM | GM | AM | CFM | CTX | CAZ | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S (26) | R (74) | P | S (57) | R (43) | P | S (47) | R (53) | P | S (81) | R (19) | P | S (8) | R (92) | P | S (29) | R (71) | P | S (35) | R (65) | P- | S (45) | R (55) | P | |
| fimH (95) | 26 | 69 | 0.87 | 55 | 40 | 1 | 45 | 50 | 1 | 77 | 18 | 1 | 7 | 88 | 1 | 29 | 66 | 0.87 | 35 | 60 | 0.88 | 44 | 51 | 0.88 |
| Sfa (81) | 23 | 58 | 0.73 | 47 | 34 | 1 | 38 | 43 | 1 | 67 | 14 | 0.84 | 7 | 74 | 1 | 25 | 56 | 0.87 | 30 | 51 | 0.87 | 38 | 43 | 0.88 |
| papC (57) | 17 | 40 | 0.71 | 29 | 28 | 0.5 | 29 | 28 | 0.7 | 47 | 10 | 1 | 6 | 51 | 0.57 | 21 | 36 | 0.37 | 25 | 32 | 0.30 | 31 | 26 | 0.31 |
| foc (16) | 4 | 12 | 1 | 10 | 6 | 0.78 | 9 | 7 | 0.59 | 13 | 3 | 1 | 2 | 14 | 0.62 | 7 | 9 | 0.25 | 7 | 9 | 0.57 | 10 | 6 | 0.28 |
| afa (12) | 5 | 7 | 0.03 | 11 | 1 | 0.15 | 7 | 5 | 0.54 | 11 | 1 | 0.68 | 0 | 12 | 0.59 | 3 | 9 | 1 | 4 | 8 | 1 | 7 | 5 | 0.54 |
| papGI (16) | 4 | 12 | 1 | 8 | 8 | 0.6 | 10 | 6 | 0.2 | 15 | 1 | 0.2 | 1 | 15 | 1 | 6 | 10 | 0.5 | 5 | 11 | 1 | 6 | 10 | 0.7 |
| papGII (34) | 10 | 24 | 0.8 | 19 | 15 | 1 | 18 | 16 | 0.5 | 28 | 6 | 1 | 3 | 31 | 1 | 12 | 22 | 0.5 | 20 | 14 | 0.01 | 15 | 19 | 1 |
| papGIII (4) | 2 | 2 | 0.2 | 2 | 2 | 1 | 2 | 2 | 1 | 3 | 1 | 0.5 | 0 | 4 | 1 | 2 | 2 | 0.5 | 2 | 2 | 0.6 | 2 | 2 | 1 |
4.2. Adhesin-Encoding Genes
The rate of the studied virulence genes is presented in Table 2. Regarding all studied adhesin factors, the fimH gene was the most common adhesin gene and was identified in 95% of the UPEC isolates followed by sfa (81%), papC (57%), papGII (34%), foc (16%), papGI (16%), afa (12%) and papGIII (4%) genes. The carriage of adhesin-encoding genes in ciprofloxacin-susceptible and -resistant isolates is also summarized in Table 2. Among the adhesin-encoding genes examined, the prevalence of fimH (55 vs. 40), sfa (47 vs. 34), papC (29 vs. 28), foc (10 vs. 6), afa (11 vs. 1), papGI (8 vs. 8), papGII (19 vs. 15), and papGIII (2 vs. 2) was higher in susceptible isolates compared with resistant isolates. As far as adhesin genes are concerned, fimH, sfa, papC, papGI, and papGII were found in a large number of isolates of both ciprofloxacin susceptible and resistant isolates, whereas afa was almost exclusively found in the ciprofloxacin susceptible isolates (11 vs. 1). Almost all of ciprofloxacin-susceptible/resistance UPEC isolates were positive for adhesin production and there was no significant discrimination between ciprofloxacin-susceptible and -resistant isolates (Table 2).
All the ciprofloxacin susceptible and resistant studied isolates exhibited 20 and 21 adhesin genes patterns, corresponding to EC, respectively, of which the following gene associations occurred more frequently in ciprofloxacin susceptible isolates: fimH, sfa, papC, foc, afa, and papGII (1 isolate), fimH, sfa, papC and foc (3 isolate), and EC6 was identified by the presence of the fimH, sfa, papC and papGII only, and was the most prominent pattern found in 11 isolates (Table 3). Among 43 ciprofloxacin-resistant isolates: fimH, sfa, papC and foc (1 isolate), fimH, sfa, papC, papGI, and papGII (4 isolates) and fimH, sfa (11 isolates), fimH, sfa, and papC (5 isolates) were the most noted pattern (Table 4). In addition, the fimH, sfa, and papC genes were more detected in the ciprofloxacin-susceptible isolates (31 vs. 17).
| Pattern | fimH | sfa | papC | foc | afa | papGI | papGII | papGIII | Number of Strains |
|---|---|---|---|---|---|---|---|---|---|
| EC1 | + | + | + | + | + | - | + | - | 1 |
| EC2 | + | + | + | + | - | - | - | - | 3 |
| EC3 | + | + | + | + | - | + | + | - | 1 |
| EC4 | + | + | + | - | - | - | - | - | 6 |
| EC5 | + | + | + | - | - | + | + | - | 3 |
| EC6 | + | + | + | - | - | - | + | - | 11 |
| EC7 | + | + | + | - | - | + | - | - | 1 |
| EC8 | + | + | + | - | - | - | - | + | 3 |
| EC9 | + | + | - | - | - | - | - | - | 8 |
| EC10 | + | - | - | - | - | - | - | - | 6 |
| EC11 | + | - | + | + | - | - | + | - | 1 |
| EC12 | + | + | + | - | + | + | - | - | 1 |
| EC13 | + | + | + | - | + | - | + | - | 1 |
| EC14 | + | + | - | + | - | - | - | - | 1 |
| EC15 | + | + | - | + | - | + | - | - | 1 |
| EC16 | + | - | + | - | - | - | - | - | 1 |
| EC17 | - | + | - | - | + | - | - | - | 1 |
| EC18 | + | + | - | - | + | - | - | - | 5 |
| EC19 | + | + | - | - | - | + | - | - | 1 |
| EC20 | + | - | - | + | - | - | - | - | 1 |
| Total | 56 | 48 | 33 | 9 | 9 | 8 | 18 | 3 | 57 |
Adhesin Gene Profile Pattern Among Ciprofloxacin-Resistant Escherichia coli Isolates
| Pattern | fimH | sfa | papC | foc | afa | papGI | papGII | papGIII | Number of Strains |
|---|---|---|---|---|---|---|---|---|---|
| EC1 | + | + | + | + | - | - | - | - | 1 |
| EC2 | + | + | + | + | - | - | + | - | 2 |
| EC3 | + | + | + | + | - | + | + | - | 2 |
| EC4 | + | + | + | - | - | + | + | - | 4 |
| EC5 | + | + | + | - | - | - | + | - | 3 |
| EC6 | + | + | + | - | - | - | - | - | 5 |
| EC7 | + | + | - | - | - | - | - | - | 11 |
| EC8 | + | + | - | - | - | - | + | - | 1 |
| EC9 | + | - | - | - | - | - | - | - | 2 |
| EC10 | - | + | + | - | - | + | + | - | 1 |
| EC11 | - | + | + | - | - | - | - | - | 1 |
| EC12 | + | + | - | + | - | - | - | - | 1 |
| EC13 | + | - | + | - | - | + | + | - | 1 |
| EC14 | + | - | + | - | - | - | - | + | 1 |
| EC15 | + | - | + | - | - | - | + | - | 1 |
| EC16 | + | - | + | - | + | - | + | - | 1 |
| EC17 | + | - | - | + | - | - | - | - | 1 |
| EC18 | + | - | - | - | + | - | - | - | 1 |
| EC19 | - | + | - | - | + | - | - | - | 1 |
| EC20 | + | - | + | - | - | - | - | - | 1 |
| EC21 | - | - | - | - | - | - | - | - | 1 |
| Total | 39 | 33 | 24 | 7 | 3 | 8 | 16 | 1 | 43 |
4.3. Serotype Structure
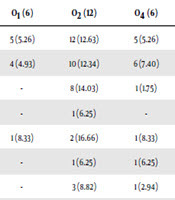
Of 100 UPEC isolates, 73 of the isolates belonged to the assessed 12 O-serogroups. Twenty-seven isolates could not be typed as they were either rough or non-typeable. The sero-groups O2, O6, O18, O157, O75, O4, O1, O16, O25, O7, and O12 were present in 16.43%, 16.43%, 13.69%, 10.95%, 9.58%, 8.21%, 8.21%, 6.84%, 4.10%, 2.73%, and 2.73% of UPEC isolates, respectively. There was no positive isolate for O15. Out of 73 O-serotyped UPEC isolates 69 (95%) and 59 (81%) were positive for fimH and sfa genes which were the most two prevalent genetic markers. In total, 73%, 73%, 72%, 63%, 92%, 81%, 79%, and 100% of fimH, sfa, papC, foc, afa, papGI, papGII, and papGIII were found in all O-serotyped isolates (Table 5). All of the detected adhesin-encoding genes were present alone or in combination with each other. Our results showed that sfa, fimH, papC, foc, afa, papGI, and papGII were found in 100%, 90%, 80%, 50%, 10%, 40%, and 60% of O18 positive isolates, respectively. Of various papG alleles, allele III were more prevalent in O6 isolates (3 out of 4 isolates) and allele II was harbored by 18%, 15%, 12%, 12%, 9%, 9%, 3%, and 3% of O18, O6, O75, O16, O157, O2, O7, and O4 positive UPEC isolates, respectively (Table 5). Our data found that all O1, O12, and O25 positive isolates were negative for the three alleles of papG. The genetic marker for papC was found in 41 (56.16%) isolates belonging to different O-serotype, which was the third most prevalent adhesin gene.
Common Virulence Factors, Antibiotic Resistance, and Genetic Among O-Serotype Escherichia coli Isolates of Human
| O 1 (6) | O 2 (12) | O 4 (6) | O 6 (12) | O 7 (2) | O 12 (2) | O 16 (50) | O 18 (10) | O 25 (3) | O 75 (7) | O 157 (8) | Total (73) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genetic | ||||||||||||
| fimH (95) | 5 (5.26) | 12 (12.63) | 5 (5.26) | 12 (12.63) | 2 (2.10) | 2 (2.10) | 5 (5.26) | 9 (9.47) | 2 (2.10) | 7 (7.36) | 8 (8.42) | 69 (73) |
| Sfa (81) | 4 (4.93) | 10 (12.34) | 6 (7.40) | 7 (8.64) | 2 (2.46) | 2 (2.46) | 4 (4.93) | 10 (12.34) | 1 (1.23) | 6 (7.40) | 7 (8.64) | 59 (73) |
| papC (57) | - | 8 (14.03) | 1 (1.75) | 8 (14.03) | 1 (1.75) | - | 5 (8.77) | 8 (14.03) | 1 (1.75) | 7 (12.28) | 2 (3.50) | 41 (72) |
| foc (16) | - | 1 (6.25) | - | 3 (18.75) | - | - | - | 5 (31.25) | - | 1 (6.25) | - | 10 (63) |
| afa (12) | 1 (8.33) | 2 (16.66) | 1 (8.33) | 2 (16.66) | - | 1 (8.33) | 2 (16.66) | 1 (8.33) | - | 1 (8.33) | - | 11 (92) |
| papGI (16) | - | 1 (6.25) | 1 (6.25) | 1 (6.25) | 1 (6.25) | - | 2 (12.5) | 4 (25) | - | 1 (6.25) | 2 (12.5) | 13 (81) |
| papGII (34) | - | 3 (8.82) | 1 (2.94) | 5 (14.70) | 1 (2.94) | - | 4 (11.76) | 6 (17.64) | - | 4 (11.76) | 3 (8.82) | 27 (79) |
| papGIII (4) | - | - | - | 3 (75) | - | - | 1 (25) | - | - | - | 4 (100) | |
| Antibiotic resistance | ||||||||||||
| AZM | ||||||||||||
| S | 5 | 7 | 4 | 4 | 1 | 2 | 4 | 6 | 2 | 2 | 5 | |
| R | 0 | 5 | 2 | 8 | 1 | 0 | 1 | 4 | 1 | 5 | 3 | |
| AM | ||||||||||||
| S | 5 | 8 | 6 | 10 | 1 | 2 | 5 | 10 | 3 | 7 | 8 | |
| R | 0 | 4 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| CZ | ||||||||||||
| S | 5 | 8 | 4 | 6 | 1 | 2 | 4 | 9 | 3 | 6 | 8 | |
| R | 0 | 4 | 2 | 5 | 1 | 2 | 1 | 1 | 0 | 1 | 0 | |
| CFM | ||||||||||||
| S | 4 | 7 | 4 | 5 | 1 | 2 | 4 | 8 | 3 | 6 | 7 | |
| R | 1 | 5 | 2 | 6 | 1 | 2 | 1 | 2 | 0 | 1 | 1 | |
| CTX | ||||||||||||
| S | 5 | 5 | 4 | 5 | 1 | 2 | 4 | 8 | 2 | 5 | 7 | |
| R | 0 | 7 | 2 | 6 | 1 | 0 | 1 | 2 | 1 | 2 | 0 | |
| CAZ | ||||||||||||
| S | 3 | 3 | 4 | 3 | 1 | 2 | 3 | 7 | 1 | 6 | 6 | |
| R | 2 | 9 | 2 | 8 | 1 | 0 | 2 | 3 | 2 | 1 | 2 | |
| GM | ||||||||||||
| S | 2 | 2 | 3 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| R | 3 | 10 | 0 | 7 | 2 | 2 | 5 | 10 | 3 | 7 | 7 | |
| CP | ||||||||||||
| S | 1 | 2 | 4 | 4 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | |
| R | 4 | 9 | 2 | 7 | 2 | 1 | 4 | 9 | 3 | 7 | 8 |
5. Discussion
A better knowledge of the frequency of genes coding for fimbrial adhesive systems such as fimH, papC, and papG alleles of UPEC strains, especially in ciprofloxacin-susceptible/resistant UPEC isolates allows the scientists to pursue the pattern of pathogenicity of strains causing the UTIs (17, 18). Since ciprofloxacin is currently used in protocols to treat UTIs, we feel it is appropriate to have a stronger knowledge of the interaction between the ciprofloxacin-resistance and the modest changes in the prevalence of genes involved in pathogenicity, especially adhesin encoding genes. Therefore, as ciprofloxacin use in UTIs, the frequency of ciprofloxacin-resistant is increasing among clinical isolates (19), clinicians may begin to see a change in the patterns of urinary tract disease accompanied by modifications in the prevalence of E. coli virulence determinants. Ciprofloxacin usually prevents the synthesis of bacterial DNA through the inhibition of two DNA gyrase enzymes and topoisomerase, which are essential for bacterial viability (20). However, the frequency of genes involved in virulence in both ciprofloxacin-sensitive and -resistant UPEC strains is not obvious.
It has been proposed that ciprofloxacin-resistant bacteria may lose some virulence genes due to decreased efficiency of gyrase and topoisomerase (21). In contrast, the development of resistance to some beta-lactam antibiotics, such as ampicillin, correlates with adhesin-encoding genes, which play an important role in bacterial colonization (22). In this study, 57 and 43 isolates were susceptible and resistant to ciprofloxacin, respectively that is consistent with the finding of Alishahi et al. in Estahban-Iran (23). In our study, as we expected, almost all the isolates (95%) harbored the fimH gene were consistent with some previous reports (24, 25). Conversely, in agreement with other published data in Iran (26), the prevalence of fimH was reported 64 percent. Moreover, the fimH gene was highly conserved (55%, 40%) in both ciprofloxacin-susceptible and -resistant isolates, emphasizing its important role during urinary tract colonization (27, 28). In addition, 34% of the isolates had papG II that is responsible for encoding PapG adhesion on the tips of P fimbriae. In other studies, it was realized that various classes of PapG adhesion were dominant in the E. coli strains isolated from UTIs (6, 29). In the current study, the frequency of papGI and papGIII were 16% and 4%, respectively. It was presented that class II papG allele was contributed to pyelonephritis cases, while papGIII was initially associated with UTIs in dogs and cats (6).
The sfa was the second most predominant adhesin gene (81%) in our isolates that was in agreement with a research carried out in Shiraz (Fars province, Iran) with a frequency of 79.4% (29). In previous reports, the prevalence of the sfa was conversely less than 30% (7). Nevertheless, the exact role of S-fimbriae is not clarified; therefore, the distribution of bacterium within the host tissue depends on this adhesin marker (29). In our isolates, the papC was the third most prevalent adhesin gene (57%) that was similar to a study conducted in Jahrom (Fars province, Iran) (30). The PapC usher protein was essential for the fimbriae P biogenesis regulation that was encoded by the papC gene (31) which was associated with pyelonephritis. Therefore, a high percentage of over 50%, including both ciprofloxacin-resistant (29%) and -susceptibile (28%) UPEC isolates collected from the Zabol (Sistan and Balouchestan province, Iran) population, has great potential to colonize kidneys and generate pyelonephritis (31). In the present study, the frequency of afa was 12% that might influence the development of chronic nephritis.
In addition, there is no significant correlation between the presence of adhesin-encoding genes in ciprofloxacin-resistant and -susceptible UPEC isolates, although the afa gene is higher in ciprofloxacin-susceptible isolates genes (11 vs 1) that is able to be linked to cystitits cases. This result is in accordance with the previous report by Lloyd et al. (32). In this research, it was shown that ciprofloxacin-resistant UPEC isolates presented a lower prevalence of fimbrial genes (fimH+, sfa+, papC+) compared to ciprofloxacin-susceptible ones (31 vs 17). As the same as the previous results (33), the outcomes declared that fimbrial genes had a lower prevalence in UPEC resistant extended-spectrum cephalosporins rather than susceptible isolates. A possible description was that the virulence genes like the resistance genes could be harbored on conjugative plasmids; then the incompatible resistance-encoding plasmids were outcompeting fimbrial factor encoding plasmids (33).
It was discovered that the acquisition of antibiotic resistance might lead to alterations in phenotypic and physiological properties that were referred to as “biological fitness cost”. The biological fitness cost on antibiotic resistance generally causes to decrease growth rates. More unfavorable phenotypic changes, including poor fimbrial expression, were presented in an ampicillin-resistant mutant Acinetobacter spp. strain DR1A in comparison to the wild type strain DR1 (34). Therefore, the declined fimbrial genes and adherence ability in ciprofloxacin-resistant UPEC might also be a reason for fitness trade-off for the quinolones to escape antibiotics exposure. More research is needed to explain the lower prevalence of fimbrial genes among resistant UPEC isolates. In this study, 73% of the UPEC isolates were included in the eleven O-serotype (O2, O6, O18, O157, O75, O4, O1, O16, O25, O7, and O12), which O2 and O6 were the most predominant (16.43%). Similar results have been reported recently too (15, 35).
The most usual antibacterial drugs in UTI’s treatment are trimethoprim-sulfamethoxazole, cephalosporins and semi-synthetic penicillins alone or with beta-lactamase inhibitors and quinolones (36). In the present study, no correlation was seen between the type of O antigen and antibiotic resistance. Few other studies have been conducted for the evaluation of the correlation between adhesin-encoding genes, antibiotic resistance, and O antigen expression (37, 38).
5.1. Conclusions
Altogether, E. coli that causes UTI in different patients varies in it’s their pathogenic capability and susceptibility to antimicrobial drugs and O-serotype profile. Developing guidelines for the management of UTI should be considered. Periodical surveys and formulation of antibiotic consumption policy are required to control the transmission and acquisition of antibiotic resistance.
This is the first report of the E. coli serotyping in the patients with UTI from southeast of Iran and their relation to antibiotic resistance and adhesin-encoding genes. Further research is necessary for a better understanding of the interaction between different virulence factors at the molecular level as well as the majority of the UPEC isolates that simultaneously express several VFs. Consequently, these results reinforce the international knowledge about antimicrobial resistance and the high rate of adhesin-encoding genes that encourage society to be aware of the proper use of antimicrobials.
Acknowledgements
References
-
1.
Foxman B. Epidemiology of urinary tract infections: Incidence, morbidity, and economic costs. Dis Mon. 2003;49(2):53-70. [PubMed ID: 12601337]. https://doi.org/10.1067/mda.2003.7.
-
2.
Dalhoff A. Global fluoroquinolone resistance epidemiology and implictions for clinical use. Interdiscip Perspect Infect Dis. 2012;2012:976273. [PubMed ID: 23097666]. [PubMed Central ID: PMC3477668]. https://doi.org/10.1155/2012/976273.
-
3.
Dalhoff A. Resistance surveillance studies: A multifaceted problem--the fluoroquinolone example. Infection. 2012;40(3):239-62. [PubMed ID: 22460782]. https://doi.org/10.1007/s15010-012-0257-2.
-
4.
Gagliotti C, Nobilio L, Moro ML; Emilia-Romagna Antibiotic Resistance Study Group. Emergence of ciprofloxacin resistance in Escherichia coli isolates from outpatient urine samples. Clin Microbiol Infect. 2007;13(3):328-31. [PubMed ID: 17391392]. https://doi.org/10.1111/j.1469-0691.2006.01615.x.
-
5.
Mavroidi A, Miriagou V, Liakopoulos A, Tzelepi E, Stefos A, Dalekos GN, et al. Ciprofloxacin-resistant Escherichia coli in Central Greece: Mechanisms of resistance and molecular identification. BMC Infect Dis. 2012;12:371. [PubMed ID: 23259844]. [PubMed Central ID: PMC3548683]. https://doi.org/10.1186/1471-2334-12-371.
-
6.
Qin X, Hu F, Wu S, Ye X, Zhu D, Zhang Y, et al. Comparison of adhesin genes and antimicrobial susceptibilities between uropathogenic and intestinal commensal Escherichia coli strains. PLoS One. 2013;8(4). e61169. [PubMed ID: 23593422]. [PubMed Central ID: PMC3621879]. https://doi.org/10.1371/journal.pone.0061169.
-
7.
Boroumand M, Sharifi A, Manzouri L, Khoramrooz SS, Khosravani SA. Evaluation of pap and sfa genes relative frequency P and S fimbriae encoding of uropathogenic Escherichia coli isolated from hospitals and medical laboratories; Yasuj City, Southwest Iran. Iran Red Crescent Med J. 2019;21(8). https://doi.org/10.5812/ircmj.89499.
-
8.
Ibrahim ME, Bilal NE, Hamid ME. Increased multi-drug resistant Escherichia coli from hospitals in Khartoum state, Sudan. Afr Health Sci. 2012;12(3):368-75. [PubMed ID: 23382754]. [PubMed Central ID: PMC3557680].
-
9.
Abdel-Rahim MH, El-Badawy O, Hadiya S, Daef EA, Suh SJ, Boothe DM, et al. Patterns of fluoroquinolone resistance in Enterobacteriaceae isolated from the Assiut University Hospitals, Egypt: A comparative study. Microb Drug Resist. 2019;25(4):509-19. [PubMed ID: 30457926]. https://doi.org/10.1089/mdr.2018.0249.
-
10.
Velasco M, Horcajada JP, Mensa J, Moreno-Martinez A, Vila J, Martinez JA, et al. Decreased invasive capacity of quinolone-resistant Escherichia coli in patients with urinary tract infections. Clin Infect Dis. 2001;33(10):1682-6. [PubMed ID: 11595990]. https://doi.org/10.1086/323810.
-
11.
Horcajada JP, Soto S, Gajewski A, Smithson A, Jimenez de Anta MT, Mensa J, et al. Quinolone-resistant uropathogenic Escherichia coli strains from phylogenetic group B2 have fewer virulence factors than their susceptible counterparts. J Clin Microbiol. 2005;43(6):2962-4. [PubMed ID: 15956432]. [PubMed Central ID: PMC1151912]. https://doi.org/10.1128/JCM.43.6.2962-2964.2005.
-
12.
Vila J, Simon K, Ruiz J, Horcajada JP, Velasco M, Barranco M, et al. Are quinolone-resistant uropathogenic Escherichia coli less virulent? J Infect Dis. 2002;186(7):1039-42. [PubMed ID: 12232848]. https://doi.org/10.1086/342955.
-
13.
Soto SM, Jimenez de Anta MT, Vila J. Quinolones induce partial or total loss of pathogenicity islands in uropathogenic Escherichia coli by SOS-dependent or -independent pathways, respectively. Antimicrob Agents Chemother. 2006;50(2):649-53. [PubMed ID: 16436722]. [PubMed Central ID: PMC1366871]. https://doi.org/10.1128/AAC.50.2.649-653.2006.
-
14.
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 28th ed. Wayne, PA USA: CLSI; 2018.
-
15.
Rashki A. Cervico-vaginopathogenic Escherichia coli in Iran: Serogroup distributions, virulence factors and antimicrobial resistance properties. Microb Pathog. 2014;75:29-34. [PubMed ID: 25193497]. https://doi.org/10.1016/j.micpath.2014.08.004.
-
16.
Clermont O, Johnson JR, Menard M, Denamur E. Determination of Escherichia coli O types by allele-specific polymerase chain reaction: Application to the O types involved in human septicemia. Diagn Microbiol Infect Dis. 2007;57(2):129-36. [PubMed ID: 17020797]. https://doi.org/10.1016/j.diagmicrobio.2006.08.007.
-
17.
Wiles TJ, Kulesus RR, Mulvey MA. Origins and virulence mechanisms of uropathogenic Escherichia coli. Exp Mol Pathol. 2008;85(1):11-9. [PubMed ID: 18482721]. [PubMed Central ID: PMC2595135]. https://doi.org/10.1016/j.yexmp.2008.03.007.
-
18.
Verma R, Rojas TC, Maluta RP, Leite JL, da Silva LP, Nakazato G, et al. Fimbria-encoding gene yadC has a pleiotropic effect on several biological characteristics and plays a role in avian pathogenic Escherichia coli Pathogenicity. Infect Immun. 2016;84(1):187-93. [PubMed ID: 26502907]. [PubMed Central ID: PMC4694002]. https://doi.org/10.1128/IAI.01138-15.
-
19.
Kahlmeter G, Menday P, Cars O. Non-hospital antimicrobial usage and resistance in community-acquired Escherichia coli urinary tract infection. J Antimicrob Chemother. 2003;52(6):1005-10. [PubMed ID: 14613955]. https://doi.org/10.1093/jac/dkg488.
-
20.
Oethinger M, Podglajen I, Kern WV, Levy SB. Overexpression of the marA or soxS regulatory gene in clinical topoisomerase mutants of Escherichia coli. Antimicrob Agents Chemother. 1998;42(8):2089-94. [PubMed ID: 9687412]. [PubMed Central ID: PMC105868].
-
21.
Gillespie SH, Voelker LL, Dickens A. Evolutionary barriers to quinolone resistance in Streptococcus pneumoniae. Microb Drug Resist. 2002;8(2):79-84. [PubMed ID: 12118521]. https://doi.org/10.1089/107662902760190617.
-
22.
Hart A, Nowicki BJ, Reisner B, Pawelczyk E, Goluszko P, Urvil P, et al. Ampicillin-resistant Escherichia coli in gestational pyelonephritis: Increased occurrence and association with the colonization factor Dr adhesin. J Infect Dis. 2001;183(10):1526-9. [PubMed ID: 11319690]. https://doi.org/10.1086/320196.
-
23.
Alishahi H, Eslami G, Zandi H, Vakili M. Frequency of QnrA and QnrB ciprofloxacin-resistant genes in Escherchia coli strains isolated from urinary tract infections in Estahban-Hospitals in Fars province. J Shahid Sadoughi Univ Med Sci. 2015;23(8):736-46.
-
24.
Lee JH, Subhadra B, Son YJ, Kim DH, Park HS, Kim JM, et al. Phylogenetic group distributions, virulence factors and antimicrobial resistance properties of uropathogenic Escherichia coli strains isolated from patients with urinary tract infections in South Korea. Lett Appl Microbiol. 2016;62(1):84-90. [PubMed ID: 26518617]. https://doi.org/10.1111/lam.12517.
-
25.
Dias RCB, Vieira MA, Moro AC, Ribolli DFM, Monteiro ACM, Camargo CH, et al. Characterization of Escherichia coli obtained from patients undergoing peritoneal dialysis and diagnosed with peritonitis in a Brazilian centre. J Med Microbiol. 2019;68(9):1330-40. [PubMed ID: 31347999]. https://doi.org/10.1099/jmm.0.001043.
-
26.
Ghazvini H, Taheri K, Edalati E, Sedighi M, Mirkalantari S. Virulence factors and antimicrobial resistance in uropathogenic Escherichiacoli strains isolated from cystitis and pyelonephritis. Turk J Med Sci. 2019;49(1):361-7. [PubMed ID: 30761847]. https://doi.org/10.3906/sag-1805-100.
-
27.
Mulvey MA. Adhesion and entry of uropathogenic Escherichia coli. Cell Microbiol. 2002;4(5):257-71. [PubMed ID: 12027955].
-
28.
Guyer DM, Gunther N, Mobley HL. Secreted proteins and other features specific to uropathogenic Escherichia coli. J Infect Dis. 2001;183 Suppl 1:S32-5. [PubMed ID: 11171010]. https://doi.org/10.1086/318854.
-
29.
Malekzadegan Y, Khashei R, Sedigh Ebrahim-Saraie H, Jahanabadi Z. Distribution of virulence genes and their association with antimicrobial resistance among uropathogenic Escherichia coli isolates from Iranian patients. BMC Infect Dis. 2018;18(1):572. [PubMed ID: 30442101]. [PubMed Central ID: PMC6238375]. https://doi.org/10.1186/s12879-018-3467-0.
-
30.
Asadi S, Kargar M, Solhjoo K, Najafi A, Ghorbani-Dalini S. The association of virulence determinants of uropathogenic Escherichia coli with antibiotic resistance. Jundishapur J Microbiol. 2014;7(5). e9936. [PubMed ID: 25147722]. [PubMed Central ID: PMC4138644]. https://doi.org/10.5812/jjm.9936.
-
31.
Antao EM, Wieler LH, Ewers C. Adhesive threads of extraintestinal pathogenic Escherichia coli. Gut Pathog. 2009;1(1):22. [PubMed ID: 20003270]. [PubMed Central ID: PMC2797515]. https://doi.org/10.1186/1757-4749-1-22.
-
32.
Lloyd AL, Rasko DA, Mobley HL. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol. 2007;189(9):3532-46. [PubMed ID: 17351047]. [PubMed Central ID: PMC1855899]. https://doi.org/10.1128/JB.01744-06.
-
33.
Zhao L, Chen X, Zhu X, Yang W, Dong L, Xu X, et al. Prevalence of virulence factors and antimicrobial resistance of uropathogenic Escherichia coli in Jiangsu province (China). Urology. 2009;74(3):702-7. [PubMed ID: 19362346]. https://doi.org/10.1016/j.urology.2009.01.042.
-
34.
Kang YS, Park W. Trade-off between antibiotic resistance and biological fitness in Acinetobacter sp. strain DR1. Environ Microbiol. 2010;12(5):1304-18. [PubMed ID: 20192959]. https://doi.org/10.1111/j.1462-2920.2010.02175.x.
-
35.
Ananias M, Yano T. Serogroups and virulence genotypes of Escherichia coli isolated from patients with sepsis. Braz J Med Biol Res. 2008;41(10):877-83. [PubMed ID: 19030710]. https://doi.org/10.1590/s0100-879x2008001000008.
-
36.
Arslan H, Azap OK, Ergonul O, Timurkaynak F; Urinary Tract Infection Study Group. Risk factors for ciprofloxacin resistance among Escherichia coli strains isolated from community-acquired urinary tract infections in Turkey. J Antimicrob Chemother. 2005;56(5):914-8. [PubMed ID: 16174685]. https://doi.org/10.1093/jac/dki344.
-
37.
Tarr PI, Clausen CR, Whittam TS, Wilson RA. Antibiotic resistance and O antigen expression in Escherichia coli. J Infect Dis. 1998;177(6):1774-5. [PubMed ID: 9607874]. https://doi.org/10.1086/517420.
-
38.
Hartley CL, Howe K, Linton AH, Linton KB, Richmond MH. Distribution of R plasmids among the O-antigen types of Escherichia coli isolated from human and animal sources. Antimicrob Agents Chemother. 1975;8(2):122-31. [PubMed ID: 1101815]. [PubMed Central ID: PMC429278]. https://doi.org/10.1128/aac.8.2.122.