Abstract
Background:
The eradication of multidrug-resistant Helicobacter pylori has become a major problem for medical care.Objectives:
In this study, the probiotics supernatant against drug-resistant H. pylori has been investigated.Methods:
First, the antimicrobial susceptibilities of the six H. pylori isolated from patients were determined by E-test. Second, Lactobacillus isolates with anti-H. pylori were activity isolated from healthy baby feces in order to inhibit the growth of H. pylori HP3. Furthermore, physical and chemical properties of antimicrobial fraction S18 and the viability of Lactobacillus reuteri S18 were investigated in simulated gastric fluid and simulated intestinal fluid.Results:
Multidrug-resistant H. pylori HP3 isolates showing four typical antibiotic resistances were selected for further study in which Lactobacillus isolates inhibited the growth of the H. pylori HP3 by producing lactate and acetic acid, and its anti-H. pylori activity disappeared after the neutralization of pH. Lactobacillus reuteri S18 with an inhibitory value (∼75%) were selected as a fighter against H. pylori HP3. Antimicrobial fraction S18 from L. reuteri S18 is stable in pepsin and is inactivated by trypsin, lysozyme and protease K. In the end, it was observed that the growth of H. pylori HP3 was completely suppressed by the antimicrobial fraction S18 at a working time of 8 h. Moreover, the cell viability of L. reuteri S18 was decreased by 20.8% and 62.5% after treatment in simulated gastric fluid and simulated intestinal fluid, respectively. Lactobacillus reuteri S18 can survive in the stomach (acidic environment) fighting for the H. pylori HP3, which can be suppressed in the intestine subsequently.Conclusions:
Lactobacillus reuteri S18 holds the potential to be applied to medical care against multidrug-resistant H. pylori.Keywords
Probiotics Supernatant Helicobacter pylori Inhibition Organic Acid Lactic Acid Multidrug Resistance
1. Background
Helicobacter pylori is the most important pathogen in chronic gastritis, peptic ulcers, and gastric cancer (1, 2). When the H. pylori infection occurs, it develops toward persistent chronic infection in both developed and developing countries (3). particularly, the prevalence was up to 80% - 90% in developing countries (4, 5). Helicobacter pylori infection with a high prevalence (approximately 50%) found in the human stomach has become a serious and common public health problem (6-8).
Historically, antibiotics combination has been used to eradicate the infection. However, the success of eradication therapies has declined and the side-effects are obvious in recent years, in part due to the development of drug-resistant H. pylori strains (9) caused by antibiotic overuse or misuse. Amoxicillin-resistant, clarithromycin-resistant (10, 11), and metronidazole-resistant H. pylori have been found in recent reports (12-14). Therefore, new safe and efficient therapies are urgently required.
Currently, probiotics cultures, one of the potential current therapies, have become more popular than the traditional eradication methods on H. pylori infection therapies. A large number of studies have demonstrated that several probiotics strains (such as Lactobacillus and Bifidobacterium) showed anti-H. pylori activity in vitro and in vivo (15). In addition, H. pylori eradication rates can be improved by probiotics supplementation (16). The possible mechanisms of anti-H. pylori activity of probiotics were shown as follows: decrease in H. pylori urease activity and adhesion; direct inhibition; secretion of lactic acid, bacteriocins, autolysins and suppression of H. pylori-associated IL-8 production (17).
2. Objectives
In this study, we aimed to screen probiotics, such as Lactobacillus isolates obtained from the healthy baby feces and try to reveal the mechanism of probiotics supernatant in H. pylori inhibition. Moreover, anti-H. pylori activity of cell-free supernatants (CFS) from seven probiotics strains against the multidrug-resistant H. pylori HP3 were evaluated.
3. Methods
3.1. Isolation and Drug Susceptibility Testing of Clinical Helicobacter pylori Strains
The tissues were collected from 17 positive male patients undergoing gastrointestinal endoscopy in Huangshan city People’s Hospital from December 2017 to February 2018. Helicobacter pylori infection was positive in 6 patients and H. pylori were isolated from corpus gastric biopsies of male patients who had no history of eradication treatment. Helicobacter pylori isolates were cultivated in 10% urea agar medium and the color change was observed following incubation 3 hours at 35°C under microaerophilic conditions. In order to obtain a pure culture, several small round colonies were selected and sub-cultured. Helicobacter pylori were cultured on H. Pylori Medium agar (bovine brain extract 4.0 g/L, beef heart extract 4.0 g/L, peptone 5.0 g/L, casein peptone 16.0 g/L, yeast extract 2.5 g/L, sodium chloride 5.0 g/L, glucose 2.0 g/L, disodium hydrogen phosphate 2.5 g/L, agar 13.5g/L) (Haibo Biotech. Co., Ltd. China) supplemented with 10% horse blood, 5% fetal calf serum, 5 µg/L trimetoprime, 5 µg/L amphotericin B, and 10 µg/L vancomycin. The culture was performed under microaerophilic conditions at 37°C. Six H. pylori with a positive oxidase, catalase, and urease testing were further identified by sequencing (Sangon tech, Shanghai, China). The isolated strains were stored at -80°C in 15% glycerol. According to the method of Ghorbani et al. (18), the isolated H. pylori strains were identified by PCR based on 16S rDNA sequences, with the primers of Forward-TTTATGGAGAGTTTGATCCTGGCTC and Reverse-AAGGAGGTGATCCAGCCGCAGGTTC.
Epsilometer test (E-test) was used to determine the minimum inhibitory concentrations (MICs) of amoxicillin (AMX), ciprofloxacin (CIP), clarithromycin (CLA), metronidazole (MTZ), tetracycline (TET), and Levofloxacin (LEV). Columbia blood agar base (Hopebiol, China) was used as culture media. The corresponding antibiotic was added in the plate and incubated for 72 hours at 37°C under microaerophilic conditions. Strains were considered resistant when the MIC was > 0.5 mg/L for amoxicillin, 1.0 mg/L for ciprofloxacin, > 0.5 mg/L for clarithromycin, > 8 mg/L for metronidazole, > 4.0 mg/L for tetracycline and > 1.0 mg/L for Levoflxacin according to the method of Shi et al. (19).
3.2. Screening of Functional Probiotics Against Helicobacter pylori
The guardian of the healthy baby (stool donors) provided informed consent. Lactobacillus was isolated from healthy baby feces according to the method of Oh et al. (20) with minor changes. In brief, the healthy baby feces samples (2 g) were weighed and homogenized for 2 minutes containing 50 mL of peptone water. The homogenized samples were spread on the surface of MRS (de Man, Rogosa and Sharpe) agar plates and incubated at 37°C for 72 hours. The pure cultures of the isolates were obtained by selected and sub-cultured several times and preserved in MRS broth containing 30% (v/v) glycerol at -80°C. Lactobacillus isolates were further identified by sequencing (Sangon tech, Shanghai, China).
Cell-free supernatant of the candidate probiotics was concentrated to 3 mg/mL by an FD-1A-100 lyophilizer (Shanghai, China). Anti-H. pylori activity of the Lactobacillus isolates cell-free supernatant was tested by the detection of the changes in absorbance using a UV-VIS spectrophotometer (Lab-Tech, China). Inhibition rate (%) = (A0 - A1) / A0 × 100%, where A0 is the initial absorbance without treatment, A1 is the initial absorbance treated with cell-free supernatant of the probiotics for 30 minutes. The spot-on-lawn method. Five µL of L. candidates’ overnight culture (1 × 107 CFU/mL) was spotted on the surface of MRS agar plates and incubated overnight at 37°C. Next, H. pylori isolates from overnight culture (1 × 107 CFU/mL) were added into the 0.7% soft agar (contained 1:1 mixture of BHI and MRS). The mixture was overlaid on the MRS agar plate containing the spots of L. candidates. The zone free of bacterial growth observed around the spots was measured in millimeters.
3.3. Production and Purification of an Antimicrobial Fraction
The cell-free supernatant of Lactobacillus S18 was purified by (NH4)2SO4 precipitation and ultrafiltration, successively. The pure fractions were firstly separated by preparative high-performance liquid chromatography (Shimadzu, Japan) with a reversed-phase (RP) column. The mobile phases consisted of 0.2% TFA in water as fluid phase A and 100% acetonitrile as fluid phase B. The flow rate was 1.0 mL/min and the absorbance was recorded at 215 nm. Fractions were collected and tested for antimicrobial activity. The active fraction was concentrated and lyophilized. Antimicrobial activity of fraction S18 was expressed as AU/mL. One AU was defined as the reciprocal of the highest dilution showing a clear zone of H. Pylori HP3 growth inhibition according to the report (21).
3.4. Effect of Enzymes on Antimicrobial Fraction Activity
Lactobacillus strains were grown in MRS broth at 30°C for 24 hours, the cells were harvested by centrifugation (5000 rpm, 20 minutes, 4°C), and the pH-neutralized supernatants was adjusted to pH = 7.0 with 2.0 mol/L NaOH. Enzymes were purchased from Sigma (China). According to the method of Ivanova et al. (22), cell-free pH-neutralized supernatant (1 mL) was incubated for one hour in the presence of 0.5 mg/mL of proteinase K, pepsin and trypsin, α-amylase and lysozyme, respectively.
3.5. Assays for Helicobacter pylori Adhesion and Invasion to AGS Cells
Assays were done for H. pylori adhesion to (human gastric cancer cells) AGS cells. The live Lactobacillus strains in the suspension were added directly to the cell culture for 20 minutes, then inoculated with H. pylori HP3 for 5 hours. To determine the number of cell-adhesion bacteria, the infected cells were washed three times to remove unbound H. pylori and the AGS cells lysed with distilled water for 20 minutes. The lysates were then diluted in PBS and cultured on Brucella blood agar plates. The viable CFUs were counted 4 days later. Assays for H. pylori invasion to AGS Cells. To determine the number of intracellular H. pylori HP3, the infected AGS cells were washed three times with PBS. In order to remove extracellular bacteria, the infected AGS cells were incubated with 200 µg/mL gentamicin for 4 hours at 37°C. The AGS cells were lysed with distilled water for 20 minutes. The lysates were then diluted in PBS and cultured on Brucella blood agar plates. The viable CFUs were counted 4 days later. The controls, containing H. pylori HP3-infected AGS cells without Lactobacillus strains were used to define as 100%.
3.6. Lactic Acid and Acetic Acid Assays and Bacterial Killing Assay
High-performance liquid chromatography (HPLC-Agilent 1100, Germany) equipped with HPX-87H (Bio-Rad Laboratories, USA) and a refractive index (IR) detector was used to determine the concentration of lactic acid and acetic acid with a column temperature of 60°C and a flux of 0.5 mL/min. The mobile phase was 0.5 mM H2SO4. Samples were diluted with of mobile phase and filtered using a 0.22 µm membrane. The activity of antimicrobial fraction S18 against H. pylori HP3 was evaluated by measuring the reduction in the numbers of CFU/mL according to the method of Urrutia-Baca et al. (23). PBS was the phosphate-buffered saline (pH = 6.0). The working concentration of the antimicrobial fraction S18 and lactic acid was 17.6 mg/mL. The concentration of the antimicrobial fraction S18 was measured by the Bradford method (24).
4. Results
4.1. Antimicrobial Susceptibilities of H. pylori Isolates
The antimicrobial susceptibilities of the six H. pylori isolates were determined by E-test in which all isolates were susceptible to AMX. However, five isolates were resistant to metronidazole, with the highest resistant rate of 83.3% (Table 1). In addition, the drug-resistant rate of CIP, CLA, TET, and LEV were 33.3%, 33.3%, 16.7%, and 33.3%, respectively. In total, H. pylori HP3 and H. pylori HP5 isolates with more than three antibiotics resistance were confirmed as multidrug-resistant (MDR). In which, H. pylori HP3 isolate, showing four antibiotics resistance was selected for further study.
In vitro, Antibiotic Susceptibility Profile of Helicobacter pylori Isolates Determined by E-Test
| Strains | MIC, mg/L | Description | |||||
|---|---|---|---|---|---|---|---|
| CIP | MTZ | AMX | CLA | TET | LEV | Resistant Antibiotics | |
| Helicobacter pylori HP1 | < 0.5 | > 128 | 0.50 | 0.5 | 1 | 0.5 | MTZ |
| H. pylori HP2 | 32 | 8 | 0.12 | 0.25 | 2 | 16 | CIP + LEV |
| H. pylori HP3 | 0.5 | > 128 | 0.24 | > 32 | 8 | 32 | MTZ + LEV + CLA + TET |
| H. pylori HP4 | 1 | > 128 | 0.12 | 0.25 | 2 | 0.5 | MTZ |
| H. pylori HP5 | 16 | > 128 | 0.12 | 32 | 1 | 1.0 | MTZ + CIP + CLA |
| H. pylori HP6 | 0.5 | > 128 | 0.12 | 1.0 | 2 | 0.25 | MTZ |
4.2. Screening of Functional Probiotic Strains Against Helicobacter pylori
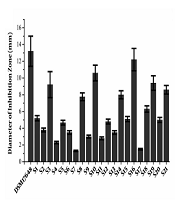
Sixty-five strains isolated from healthy baby feces were tested for their anti-H. pylori activity by the spot-on-lawn method. Thirty-six clones were obtained from MRS plate agar (pH = 2.0) and their anti-H. pylori activity has been tested. Twenty-one strains exhibited different levels of H. pylori inhibition in the range of 1.3 to 12.2 mm were shown in Figure 1. Afterward, the supernatant of thirteen isolates (S1, S3, S5, S8, S10, S12, S14, S15, S16, S18, S19, S20, S21) with a diameter of inhibition zone above 5.0 were selected to test the anti-H. pylori activity. As a result, the supernatant of five isolates (S3, S8, S15, S18, S20) could suppress the growth of H. pylori with inhibition rate above 60% (OD600nm) were selected for further study. By DNA sequencing, S3, S8, S15, S18, S20 isolates were identified as L. vaginalis, L. gasseri, L. fermentum, L. reuteri, L. casei, respectively.
Screening functional probiotic strains against Helicobacter pylori (A). The anti-H. pylori activity of the supernatant of five isolates (B). DSM17648 is on behalf of L. reuteri DSM17648 (the control), which has the ability to reduce the load of H. pylori and have been used in a human study. Data are represented as means ± standard deviation (SD) of triplicate determinations from three independent experiments.

Then pH values and organic acids concentrations of the Lactobacillus isolates from broth supernatant were investigated after 24 hours cultivation. The result indicated that the pH of the six isolates culture supernatant was approximately 3.5 - 4.0 (Figure 2A). Of which, S8 isolate showed poor performance on lactic acid and acetic acid production. Besides S8 isolate, the concentration of lactic acid produced by isolates was above 16 g/L. In order to confirm antimicrobial substance, cell-free supernatant, pH-neutralized supernatants were tested (Figure 2B). Interestingly, anti-H. pylori activity of S3 isolate and S20 isolate disappeared after adjusting the pH to 7.0. It seemed that the anti-H. pylori activity of S3 and S20 isolate was probably due to the organic acids. Moreover, it was observed that the anti-H. pylori activity of S8, S15, and S18 isolate supernatants still existed after adjusting the pH to 7.0. The anti-H. pylori activity of S8, S15 and S18 pH-neutralized supernatants was reduced by 35.37%, 50%, and 8.54% compared to that of natural-pH supernatants.
Here, pH and organic acid concentrations from Lactobacillus isolate culture supernatant after 24 hours is shown (A). The effect of pH-neutralized supernatants on inhibition of Helicobacter pylori growth (B). The pH was adjusted by 0.5 M NaOH. The effects of Lactobacillus isolates on MDR H. pylori HP3 (C) adhesion and (D) invasion into AGS cells (gastric epithelial cells). AGS cells were treated with the Lactobacillus (L. gasseri S8, L. fermentum S15, L. reuteri S18), followed by infection with H. pylori HP3 for 6 hours. These values are means of triplicate experiments. Data are represented as means ± SD.

The growth of H. pylori was inhibited by S18 strain with the inhibition of 75%. Lactobacillus inhibited the initial step of H. pylori adhesion and the invasion of AGS cells was investigated. As compared to H. pylori infection alone (control group), L. gasseri S8, L. fermentum S15, and L. reuteri S18 exhibited significant anti-adhesion activity against H. pylori HP3 by 75%, 54% and 72%, respectively (Figure 2C). In addition, L. gasseri S8, L. fermentum S15, and L. reuteri S18 exhibited dramatic inhibitory activity against H. pylori HP3 invasion to AGS cells, with a reduction of 82%, 64%, and 93.0% (Figure 2D). The antimicrobial substance of S15 isolate was a heat-labile protein in cell-free supernatant with no anti-H. pylori activity after heat-treating (Figure 3A). Therefore, S18 strain was selected for further study.
The heat resistance (A) and pH resistance (B) of antimicrobial fraction produced by three isolates (S8, S15, and S18)

4.3. Production of the Antimicrobial Substance
In order to enhance the production of antimicrobial fraction and obtain the optimum conditions, different nitrogen and carbon sources were investigated. The result illustrated that the excellent nitrogen source and carbon sources were tryptone and glucose, respectively (Figure 4A and B). However, the metallic ion has no significant effect on improving the yield of antimicrobial fraction, and the Tween 80 as an inducing factor performed well, and anti-H. pylori activity of the broth supernatant was up to 224 AU/mL, while the Tween 80 concentration was 2.0 g/L (Figure 4C). Moreover, the results demonstrated that the highest antibacterial activity of antimicrobial fraction reached 256 AU/mL after 24 hours of incubation (Figure 4D). Unfortunately, it also illustrated that when the incubation time was beyond 24 hours, the antimicrobial fraction production was not increased along with the incubation time. The antimicrobial fraction was produced in the late logarithmic phase of the S18 isolate growth process (Figure 4D).
The effect of nitrogen (A), carbon source (B), inducing factor and metallic ion (C), and incubation time (D) on the production of antimicrobial fraction S18. Data are represented as means ± SD.

4.4. Purification of the Antimicrobial Fraction S18
The results of the purification procedure are shown in Table 2 and Figure 5. The 1 L cell-free supernatant was concentrated by (NH4)2SO4 precipitation and the crude antimicrobial fraction S18 was obtained by ultrafiltration with antimicrobial activity of 1614.35 AU/mg (Table 2). At the final step of the purification process, two single peaks were obtained (Figure 5B). The lyophilized sample of the third peak at 15.2 minutes (antimicrobial fraction S18) showed a significant antibacterial activity (9600 AU/mL), which was 37.5 times stronger than that of the 24 hours cell-free supernatant (256 AU/mL) after optimization of culture conditions. By amino acid analysis and positive ion electrospray mass, the antimicrobial fraction S18 was identified as the bacteriocin (reutericin 6), which was consistent with the report of Kabuki et al. (25).
Purification of antimicrobial fraction S18 by RP-HPLC. A, Purification of antimicrobial fraction from cell-free supernatant after Gel chromatography; B,Purification of antimicrobial fraction from cell-free supernatant after RP-HPLC.

Purification of the Antimicrobial Fraction S18
| Purification | Total Protein, mg/mL | Activity, AU/mL | Specific Activity, AU/mg | Purification, fold |
|---|---|---|---|---|
| Cell free supernatant (1L) | 35.2 | 240 | 6.82 | 1 |
| (NH4)2SO4 precipitation | 24.3 | 1520 | 62.55 | 9 |
| Ultrafiltration | 8.92 | 14400 | 1614.35 | 237 |
| RP-HPLC | 2.10 | 9600 | 4571.43 | 670 |
4.5. Bactericidal Activity of Antimicrobial Fraction S18
The effects of enzymes on the activity of antimicrobial fraction S18 were investigated. The result showed that the antimicrobial fraction S18 maintained its antibacterial activity after pepsin treatment (Figure 6A). In addition, the antimicrobial activity of the antimicrobial fraction S18 significantly decreased by 80% and 50% after treating with trypsin and Lysozyme, respectively. It was observed that the antimicrobial activity was completely inactivated by protease K after incubation 15 minutes with 95% of antimicrobial activity loss compared to the control.
The effects of enzymes on the antimicrobial fraction S18 (A); Inhibition kinetics of antimicrobial fraction S18 strain against Helicobacter pylori HP3 (B). PBS was the phosphate-buffered saline (pH = 6.0); the working concentration of the antimicrobial fraction S18 and lactic acid was 16 µg/mL and 17.6 mg/mL, respectively. The concentration of the antimicrobial fraction S18 was measured by Bradford method. Data are represented as means ± SD.

In order to analyze the contribution of acid and antimicrobial fraction S18 against the H. pylori HP3, the growth of H. pylori HP3 after the antimicrobial fraction S18 and lactic acid treatment were investigated (Figure 6B). The result indicated that the antimicrobial fraction S18 (16 µg/mL) showed high antimicrobial activity (100% inhibition rate) to inhibit the growth of H. pylori HP3 at a working time of 8 hours. Moreover, the growth of H. pylori HP3 was inhibited by incubation in lactic acid (lactic acid concentration 17.6 g/L) at a working time of 20 hours (Figure 6B). In order to assess cell viability after gastric transit, the viability of L. reuteri S18 was measured in simulated gastric fluid and simulated intestinal fluid. The results showed that the cell viability of L. reuteri S18 was decreased by 20.8% and 62.5% after treatment in simulated gastric fluid and simulated intestinal fluid, respectively (Figure 7A and B). Furthermore, in order to simulate gastric transit conditions, L. reuteri S18 was exposed to the simulated gastric fluid and simulated intestinal fluid sequentially, resulting in a cell viability loss of 46% (Figure 7C and D).
Lactobacillus reuteri S18 viability in simulated gastric fluid individually (A), Lactobacillus reuteri S18 viability in simulated intestinal fluid individually (B). The simulated human upper GI tract model system in vitro (C). Lactobacillus reuteri S18 viability as determined by spread plating after treatment in simulated gastric fluid succeeded by treatment in simulated intestinal fluid (D).

5. Discussion
Multidrug-resistant H. pylori HP3 has been isolated and identified from male patients. Lactobacillus was isolated and screened from healthy baby feces against multidrug-resistant H. pylori HP3. Many Lactobacillus isolates showed anti-H. pylori activity via producing lactate and acetic acid, and its anti-H. pylori activity would disappear after pH-neutralization. The pH-neutralized supernatants of L. reuteri S18 having 75% of inhibitory value were selected as the excellent candidate probiotic. Then antimicrobial fraction S18 has been purified from L. reuteri S18. It was observed that the growth of H. pylori HP3 was completely suppressed by the antimicrobial fraction S18 at a working time of 8 hours. Moreover, in simulated gastric fluid and simulated intestinal fluid, the cell viability of L. reuteri S18 was decreased by 20.8% and 62.5% after the treatment, respectively.
In order to eliminate the H. pylori HP3, therapies with high efficiency and no/few side-effects are urgently required. To date, probiotics with the ability to inhibit or kill H. pylori may be the best choice. It has been reported that the H. pylori showed sensitivity to L. salivarius, L. gasseri, L. delbruckii subsp. bulgaricus (26, 27). Moreover, Lactobacillus and Streptococcus have been isolated from the human stomach in previous research (28, 29). In addition, Lactobacillus has been used for the prevention and treatment of many diseases. L. rhamnosus GR-1 and L. reuteri has been used for prevention and treatment of recurrent urogenital infection (30); L. casei BL23 can reduce DMH-associated colorectal cancer via regulation of Treg and Th17 populations of T-cells (31, 32); L. rhamnosus GG in the prevention of antibiotic-associated diarrhea in children and adults (33); L. rhamnosus GG supernatant has been used for prevention acute alcohol-induced liver steatosis (34).
Lactobacillus casei CRL431 can prevent breast cancer development by modification of the cytokine profiles (35). Compared with adults, babies are very healthy, not suffering from age-related diseases (such as diabetes, obesity, autoimmune diseases, and cancer). Moreover, baby feces is easy to be collected relatively (36). Therefore, intestinal microbes of the healthy baby were selected as the main source of probiotics in the present study. In addition, in order to avoid the injury of the donor, therefore, healthy baby feces were chosen as the final source. Helicobacter pylori were found and isolated in the human stomach (37), which can survive in strong acidic conditions (pH = 0.5 - 2). Only acidophilic and acid-resistant bacteria survive in the acidic environment of the gastric lumen. Therefore, the excellent probiotics were obtained by screening acid tolerance and anti-H. pylori activity, ensuring the selected strains can handle with the gastric environment and inhibit the growth of H. pylori in the present study.
Furthermore, we observed that the antimicrobial fraction S18 produced by L. reuteri S18 is stable in the presence of pepsin and is inactivated by treating with trypsin, lysozyme, and protease K. It was indicated that antimicrobial fraction S18 may be resistant to the gastric environment and can be inactivated by trypsin or another alkaline environment in the intestine. Therefore, antimicrobial fraction S18 has the potential to be used to treat the patient suffering from H. pylori infection, which can be inactivated in the intestine. Moreover, it has been observed that the growth of H. pylori HP3 was completely suppressed by the antimicrobial fraction S18 at a working time of 8 hours. It was also indicated that the growth of multidrug-resistant H. pylori HP3 was inhibited by L. reuteri S18 via the production of antimicrobial fraction S18 mainly and lactic acid secondarily. The results showed that the cell viability of L. reuteri S18 was decreased by 20.8% and 62.5% after treatment in simulated gastric fluid and simulated intestinal fluid, respectively. It demonstrates that L. reuteri S18 can survive in the stomach (acidic environment) fighting for the H. pylori HP3, which can be suppressed in the intestine subsequently. In summary, L. reuteri S18 has the potential to be applied to medical care against multidrug-resistant H. pylori HP3. This method holds the potential to be used for screening other probiotics.
Helicobacter pylori, first isolated by Warren and Marshall in 1983 (38), is now recognized as a major cause of chronic gastritis and risk of development of gastric cancer (39, 40). However, H. pylori isolates with amoxicillin, clarithromycin, or metronidazole resistance has already emerged (41, 42). In this study, 33.3%, 33.3%, 16.7%, 33.3%, and 83.3% of the tested H. pylori were resistant to CIP, CLA, TET, LEV, and MTZ, respectively. Particularly, the prevalence of metronidazole-resistant H. pylori has increased 20% and 13.3%, comparing to the report of Tomatari et al. (64%) (43) and Mousavi et al. (70%) (44), respectively. It appears that the prevalence of multidrug-resistant H. pylori is increasing.
5.1. Conclusions
In this study, H. pylori isolated from the stomach of male patients were susceptible to AMX. However, H. pylori from salad and vegetable samples had the highest levels of resistance against AMX (67.79%), and ampicillin (61.01%) in 2014 (45). Moreover, H. pylori strains isolated from raw milk samples harbored the highest prevalence of resistance against ampicillin (82.08%), AMX (74.62%) in 2018 (46). Helicobacter pylori strains from traditional dairy products harbored the highest incidence of resistance against ampicillin (93.54%) and AMX (93.54%) (47). Therefore, we speculate that the prevalence of H. pylori infection is related to milk contaminated with H. pylori. However, additional research is required to find the relationship between H. pylori infection and daily diet (such as raw milk, cheese and cream).
References
-
1.
Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347(15):1175-86. [PubMed ID: 12374879]. https://doi.org/10.1056/NEJMra020542.
-
2.
Peek RM Jr, Blaser MJ. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2(1):28-37. [PubMed ID: 11902583]. https://doi.org/10.1038/nrc703.
-
3.
Perez-Perez GI, Sack RB, Reid R, Santosham M, Croll J, Blaser MJ. Transient and persistent Helicobacter pylori colonization in Native American children. J Clin Microbiol. 2003;41(6):2401-7. [PubMed ID: 12791856]. [PubMed Central ID: PMC156565]. https://doi.org/10.1128/jcm.41.6.2401-2407.2003.
-
4.
Jarosz M, Rychlik E, Siuba M, Respondek W, Ryzko-Skiba M, Sajor I, et al. Dietary and socio-economic factors in relation to Helicobacter pylori re-infection. World J Gastroenterol. 2009;15(9):1119-25. [PubMed ID: 19266606]. [PubMed Central ID: PMC2655195]. https://doi.org/10.3748/wjg.15.1119.
-
5.
Vale FF, Vitor JM. Transmission pathway of Helicobacter pylori: Does food play a role in rural and urban areas? Int J Food Microbiol. 2010;138(1-2):1-12. [PubMed ID: 20122750]. https://doi.org/10.1016/j.ijfoodmicro.2010.01.016.
-
6.
Go MF. Review article: Natural history and epidemiology of Helicobacter pylori infection. Aliment Pharmacol Ther. 2002;16 Suppl 1:3-15. [PubMed ID: 11849122]. https://doi.org/10.1046/j.1365-2036.2002.0160s1003.x.
-
7.
Hu Y, Wan JH, Li XY, Zhu Y, Graham DY, Lu NH. Editorial: Recurrence of Helicobacter pylori infection-still the same after all these years... Authors' reply. Aliment Pharmacol Ther. 2018;47(1):132-3. [PubMed ID: 29226407]. https://doi.org/10.1111/apt.14386.
-
8.
McMahon BJ, Bruce MG, Koch A, Goodman KJ, Tsukanov V, Mulvad G, et al. The diagnosis and treatment of Helicobacter pylori infection in Arctic regions with a high prevalence of infection: Expert Commentary. Epidemiol Infect. 2016;144(2):225-33. [PubMed ID: 26094936]. [PubMed Central ID: PMC4697284]. https://doi.org/10.1017/S0950268815001181.
-
9.
Graham DY, Fischbach L. Helicobacter pylori treatment in the era of increasing antibiotic resistance. Gut. 2010;59(8):1143-53. [PubMed ID: 20525969]. https://doi.org/10.1136/gut.2009.192757.
-
10.
Duck WM, Sobel J, Pruckler JM, Song Q, Swerdlow D, Friedman C, et al. Antimicrobial resistance incidence and risk factors among Helicobacter pylori-infected persons, United States. Emerg Infect Dis. 2004;10(6):1088-94. [PubMed ID: 15207062]. [PubMed Central ID: PMC3323181]. https://doi.org/10.3201/eid1006.030744.
-
11.
Park JY, Dunbar KB, Mitui M, Arnold CA, Lam-Himlin DM, Valasek MA, et al. Helicobacter pylori clarithromycin resistance and treatment failure are common in the USA. Dig Dis Sci. 2016;61(8):2373-80. [PubMed ID: 26923948]. https://doi.org/10.1007/s10620-016-4091-8.
-
12.
Koletzko S, Richy F, Bontems P, Crone J, Kalach N, Monteiro ML, et al. Prospective multicentre study on antibiotic resistance of Helicobacter pylori strains obtained from children living in Europe. Gut. 2006;55(12):1711-6. [PubMed ID: 16603633]. [PubMed Central ID: PMC1856474]. https://doi.org/10.1136/gut.2006.091272.
-
13.
Megraud F. Helicobacter pylori and antibiotic resistance. Gut. 2007;56(11):1502. [PubMed ID: 17938430]. [PubMed Central ID: PMC2095668]. https://doi.org/10.1136/gut.2007.132514.
-
14.
Lee SM, Kim N, Kwon YH, Nam RH, Kim JM, Park JY, et al. rdxA, frxA, and efflux pump in metronidazole-resistant Helicobacter pylori: Their relation to clinical outcomes. J Gastroenterol Hepatol. 2018;33(3):681-8. [PubMed ID: 28748532]. https://doi.org/10.1111/jgh.13906.
-
15.
Gotteland M, Brunser O, Cruchet S. Systematic review: Are probiotics useful in controlling gastric colonization by Helicobacter pylori? Aliment Pharmacol Ther. 2006;23(8):1077-86. [PubMed ID: 16611267]. https://doi.org/10.1111/j.1365-2036.2006.02868.x.
-
16.
Ruggiero P. Use of probiotics in the fight against Helicobacter pylori. World J Gastrointest Pathophysiol. 2014;5(4):384-91. [PubMed ID: 25400981]. [PubMed Central ID: PMC4231502]. https://doi.org/10.4291/wjgp.v5.i4.384.
-
17.
Hauser G, Salkic N, Vukelic K, JajacKnez A, Stimac D. Probiotics for standard triple Helicobacter pylori eradication: A randomized, double-blind, placebo-controlled trial. Medicine (Baltimore). 2015;94(17). e685. [PubMed ID: 25929897]. [PubMed Central ID: PMC4603068]. https://doi.org/10.1097/MD.0000000000000685.
-
18.
Ghorbani F, Gheisari E, Safarpoor Dehkordi F. Genotyping of vacA alleles of Helicobacter pylori strains recovered from some Iranian food items. Tropical Journal of Pharmaceutical Research. 2016;15(8). https://doi.org/10.4314/tjpr.v15i8.5.
-
19.
Shi J, Jiang Y, Zhao Y. Promising in vitro and in vivo inhibition of multidrug-resistant Helicobacter pylori by linezolid and novel oxazolidinone analogues. J Glob Antimicrob Resist. 2016;7:106-9. [PubMed ID: 27718441]. https://doi.org/10.1016/j.jgar.2016.07.016.
-
20.
Oh NS, Joung JY, Lee JY, Kim Y. Probiotic and anti-inflammatory potential of Lactobacillus rhamnosus 4B15 and Lactobacillus gasseri 4M13 isolated from infant feces. PLoS One. 2018;13(2). e0192021. [PubMed ID: 29444150]. [PubMed Central ID: PMC5812581]. https://doi.org/10.1371/journal.pone.0192021.
-
21.
van Reenen CA, Dicks LM, Chikindas ML. Isolation, purification and partial characterization of plantaricin 423, a bacteriocin produced by Lactobacillus plantarum. J Appl Microbiol. 1998;84(6):1131-7. [PubMed ID: 9717299]. https://doi.org/10.1046/j.1365-2672.1998.00451.x.
-
22.
Ivanova I, Miteva V, Stefanova T, Pantev A, Budakov I, Danova S, et al. Characterization of a bacteriocin produced by Streptococcus thermophilus 81. Int J Food Microbiol. 1998;42(3):147-58. [PubMed ID: 9728685]. https://doi.org/10.1016/s0168-1605(98)00067-1.
-
23.
Urrutia-Baca VH, Escamilla-Garcia E, de la Garza-Ramos MA, Tamez-Guerra P, Gomez-Flores R, Urbina-Rios CS. In vitro antimicrobial activity and downregulation of virulence gene expression on Helicobacter pylori by reuterin. Probiotics Antimicrob Proteins. 2018;10(2):168-75. [PubMed ID: 29103130]. https://doi.org/10.1007/s12602-017-9342-2.
-
24.
Hammond JB, Kruger NJ. The bradford method for protein quantitation. Methods Mol Biol. 1988;3:25-32. [PubMed ID: 21400151]. https://doi.org/10.1385/0-89603-126-8:25.
-
25.
Kabuki T, Saito T, Kawai Y, Uemura J, Itoh T. Production, purification and characterization of reutericin 6, a bacteriocin with lytic activity produced by Lactobacillus reuteri LA6. Int J Food Microbiol. 1997;34(2):145-56. [PubMed ID: 9039561]. https://doi.org/10.1016/s0168-1605(96)01180-4.
-
26.
Bae IH, Choi JK, Chough C, Keum SJ, Kim H, Jang SK, et al. Potent hepatitis C virus NS5A inhibitors containing a benzidine core. ACS Med Chem Lett. 2014;5(3):255-8. [PubMed ID: 24900814]. [PubMed Central ID: PMC4027583]. https://doi.org/10.1021/ml4003293.
-
27.
Daniele S, Sestito S, Pietrobono D, Giacomelli C, Chiellini G, Di Maio D, et al. Dual inhibition of PDK1 and aurora kinase a: An effective strategy to induce differentiation and apoptosis of human glioblastoma multiforme stem cells. ACS Chem Neurosci. 2017;8(1):100-14. [PubMed ID: 27797168]. https://doi.org/10.1021/acschemneuro.6b00251.
-
28.
Ryan KA, Jayaraman T, Daly P, Canchaya C, Curran S, Fang F, et al. Isolation of lactobacilli with probiotic properties from the human stomach. Lett Appl Microbiol. 2008;47(4):269-74. [PubMed ID: 19241519]. https://doi.org/10.1111/j.1472-765x.2008.02416.x.
-
29.
Delgado S, Cabrera-Rubio R, Mira A, Suarez A, Mayo B. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb Ecol. 2013;65(3):763-72. [PubMed ID: 23397369]. https://doi.org/10.1007/s00248-013-0192-5.
-
30.
Abad CL, Safdar N. The role of lactobacillus probiotics in the treatment or prevention of urogenital infections--a systematic review. J Chemother. 2009;21(3):243-52. [PubMed ID: 19567343]. https://doi.org/10.1179/joc.2009.21.3.243.
-
31.
Lenoir M, Del Carmen S, Cortes-Perez NG, Lozano-Ojalvo D, Munoz-Provencio D, Chain F, et al. Lactobacillus casei BL23 regulates Treg and Th17 T-cell populations and reduces DMH-associated colorectal cancer. J Gastroenterol. 2016;51(9):862-73. [PubMed ID: 26749362]. https://doi.org/10.1007/s00535-015-1158-9.
-
32.
Zhong L, Zhang X, Covasa M. Emerging roles of lactic acid bacteria in protection against colorectal cancer. World J Gastroenterol. 2014;20(24):7878-86. [PubMed ID: 24976724]. [PubMed Central ID: PMC4069315]. https://doi.org/10.3748/wjg.v20.i24.7878.
-
33.
Szajewska H, Kolodziej M. Systematic review with meta-analysis: Lactobacillus rhamnosus GG in the prevention of antibiotic-associated diarrhoea in children and adults. Aliment Pharmacol Ther. 2015;42(10):1149-57. [PubMed ID: 26365389]. https://doi.org/10.1111/apt.13404.
-
34.
Zhang L, Li H, Kong X, Feng W. Lactobacillus rhamnosus GG supernatant prevents acute alcohol-induced liver steatosis and injury through ER stress and autophagy-mediated signaling pathways. FASEB J. 2019;33(1_supplement):680.13.
-
35.
Utz VEM, Perdigon G, de Moreno de LeBlanc A. Milk fermented by Lactobacillus casei CRL431 modifies cytokine profiles associated to different stages of breast cancer development in mice. Benef Microbes. 2019:1-10. [PubMed ID: 31122044]. https://doi.org/10.3920/BM2019.0011.
-
36.
Nagpal R, Wang S, Ahmadi S, Hayes J, Gagliano J, Subashchandrabose S, et al. Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Sci Rep. 2018;8(1):12649. [PubMed ID: 30139941]. [PubMed Central ID: PMC6107516]. https://doi.org/10.1038/s41598-018-30114-4.
-
37.
Montecucco C, Rappuoli R. Living dangerously: How Helicobacter pylori survives in the human stomach. Nat Rev Mol Cell Biol. 2001;2(6):457-66. [PubMed ID: 11389469]. https://doi.org/10.1038/35073084.
-
38.
Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;1(8336):1273-5. [PubMed ID: 6134060]. https://doi.org/10.1016/S0140-6736(83)92719-8.
-
39.
Hamilton-Miller JM. The role of probiotics in the treatment and prevention of Helicobacter pylori infection. Int J Antimicrob Agents. 2003;22(4):360-6. [PubMed ID: 14522098]. https://doi.org/10.1016/s0924-8579(03)00153-5.
-
40.
Amieva M, Peek RM Jr. Pathobiology of Helicobacter pylori-induced gastric cancer. Gastroenterology. 2016;150(1):64-78. [PubMed ID: 26385073]. [PubMed Central ID: PMC4691563]. https://doi.org/10.1053/j.gastro.2015.09.004.
-
41.
Wegman M A, Janssen M H A, van Rantwijk F, Sheldon R A. Towards biocatalytic synthesis of β-lactam antibiotics. Adv Synth Catal. 2001;343(6-7):559-76. https://doi.org/10.1002/1615-4169(200108)343:6/7<559::Aid-adsc559>3.0.Co;2-z.
-
42.
Regnath T, Raecke O, Enninger A, Ignatius R. Increasing metronidazole and rifampicin resistance of Helicobacter pylori isolates obtained from children and adolescents between 2002 and 2015 in southwest Germany. Helicobacter. 2017;22(1). [PubMed ID: 27400262]. https://doi.org/10.1111/hel.12327.
-
43.
Tomatari FH, Mobarez AM, Amini M, Hosseini D, Abadi ATB. Helicobacter pylori resistance to metronidazole and clarithromycin in dyspeptic patients in Iran. Iran Red Crescent Med J. 2010;12:409-12.
-
44.
Mousavi S, Dehkordi FS, Rahimi E. Virulence factors and antibiotic resistance of Helicobacter pylori isolated from raw milk and unpasteurized dairy products in Iran. J Venom Anim Toxins Incl Trop Dis. 2014;20:51. [PubMed ID: 25873940]. [PubMed Central ID: PMC4396062]. https://doi.org/10.1186/1678-9199-20-51.
-
45.
Yahaghi E, Khamesipour F, Mashayekhi F, Safarpoor Dehkordi F, Sakhaei MH, Masoudimanesh M, et al. Helicobacter pylori in vegetables and salads: Genotyping and antimicrobial resistance properties. Biomed Res Int. 2014;2014:757941. [PubMed ID: 25184146]. [PubMed Central ID: PMC4145543]. https://doi.org/10.1155/2014/757941.
-
46.
Ranjbar R, Farsani FY, Dehkordi FS. Phenotypic analysis of antibiotic resistance and genotypic study of the vacA, cagA, iceA, oipA and babA genotypes of the Helicobacter pylori strains isolated from raw milk. Antimicrob Resist Infect Control. 2018;7:115. [PubMed ID: 30288255]. [PubMed Central ID: PMC6162967]. https://doi.org/10.1186/s13756-018-0409-y.
-
47.
Ranjbar R, Yadollahi Farsani F, Safarpoor Dehkordi F. Antimicrobial resistance and genotyping of vacA, cagA, and iceA alleles of the Helicobacter pylori strains isolated from traditional dairy products. J Food Saf. 2018;39(2). https://doi.org/10.1111/jfs.12594.