Abstract
Background:
Hepatitis B Virus (HBV) is a universal health challenge all around the world. Several factors like viral load, genetic characteristics, age, sex, and immune status contribute to variable clinical outcomes of HBV infection. The sequels of HBV infection vary remarkably among persons ranging from the spontaneous deletion of infection to persistent infection.Objective:
The present study aimed to evaluate the association of single nucleotide polymorphisms IL10-1082 with HBV clearance.Methods:
Sixty subjects with Chronic Hepatitis B (CHB) infection and 60 subjects who spontaneously recovered HBV were enrolled in the study. The IL-10-1082 polymorphisms were determined by Polymerase Chain Reaction with Restriction Fragment Length Polymorphism (PCR–RFLP).Results:
The clearance of HBV infection demonstrated a significant association with IL-10-1082 polymorphisms in the GG genotype (P = 0.03), while there was no association with other genotypes. Reduced risk of chronic hepatitis B infection was associated with IL-10-1082 GG (OR: 2.33, 95% CI: 1.07 - 5.09). Besides, IL-10-1082 A/G alleles did not differ clearly between the two study groups (P = 0.07)Conclusions:
The IL-10-1082 polymorphisms may be associated with a reduced risk of CHB infection and recovery after HBV infection.Keywords
Interleukin-10 Single Nucleotide Polymorphism Chronic Hepatitis B
1. Background
Hepatitis B virus (HBV) is a member of the Hepadnaviridae family, which is a public health problem all over the world (1). This virus can transmit with body secretions of the infected individual (2). Viral hepatitis is the seventh cause of death universally, responsible for 1.34 million mortalities in 2015 (3). In 2010, the mortality rate of hepatitis B was estimated at 786,000 people (4). The Middle East is part of the world with a moderate incidence of HBV in which 1 - 7% of the population suffers from chronic hepatitis B (CHB) (5). Around 1.5 million Iranians have HBV infection, showing a mild to moderate prevalence according to the World Health Organization (WHO) classification (2).
An unusual immune response of liver cells to HBV infection can influence the clinical outcome of HBV infection (6). The sequels of HBV infection vary remarkably among persons ranging from the spontaneous deletion of disease to persistent infection, which may result in hepatic failure, CHB, or hepatocellular carcinoma (7, 8). Almost 5-10% of adults who have the acute infection will progress to chronic HBV infection (9). Although the accurate mechanisms leading to different outcomes of HBV have not been illustrated, numerous researchers have started to check the genetic factors that affect the outcomes of HBV infection (10, 11). In the recent decade, various investigations have offered that polymorphisms in the cytokines genes can be related to HBV infection and may impress the progression of disease after infection (12-14).
As known, IL-10 is an anti-inflammatory cytokine and powerful immuno-modulatory molecule that is produced by macrophages and lymphoid cells (Th2 cells, T regulatory lymphocytes) (15). By inhibiting the production of pro-inflammatory cytokines (IFN-γ, TNF-α, and IL-2), it exerts its functions in the regulation of inflammatory responses (16). There is an opinion that the capacity of cytokine production depends on the genetic content, which is attributed to the polymorphism in the regulatory region or the target sequence (17). In subjects with hepatitis B infection, the heterogeneity of the IL-10 gene acts as a likely marker for defining disease phenotypes (18). Single nucleotide polymorphisms (SNPs) are the most common changes in the genome sequence and allocate approximately 90% of the variations (19).
Several SNPs in the promoter of interleukin 10 were assessed in correlation with HBV infection, like polymorphisms on the -592 site, -819 site, and -1082 site, which form three dominant haplotypes (CCA, CCG, ATA) (20). Low levels of IL-10 have been correlated with ATA haplotypes, whereas CCG and CCA haplotypes generate high and intermediate levels of IL-10, respectively (21). Therefore, the allelic variation in IL-10 might be associated with the pathogenesis and progression of HBV infection. By checking cytokine polymorphisms and immune response, healthcare workers can forecast the severity of the disease and hence propose the most impressive therapy options to the patients (22).
2. Objectives
In this study, we compared the polymorphism of the IL-10-1082 site in CHB patients and spontaneously recovered HBV patients (as controls) to understand whether gene polymorphisms in IL-10-1082 have an association with the chronicity of HBV in the Birjand population.
3. Methods
3.1. Patients and Controls
This investigation consisted of 60 patients with CHB infection and 60 subjects who spontaneously improved HBV. Samples were selected from the hepatitis comprehensive plan in Birjand during 2013 and 2014 (23, 24). The comprehensive plan consisted of 5,235 of Birjand population aged 15 to 70 years. The comprehensive plan included 786 (15%) subjects with positive hepatitis B core antibody (anti-HBc) as subjects with recovered HBV infection and 81 (1.6%) individuals with hepatitis B surface antigen (HBsAg) seropositivity as chronic HBV-infected subjects. The CHB subjects were defined as the continuous presence of hepatitis B surface antigen (HBsAg) for more than six months. The HBsAg was detected by an enzyme-linked immunosorbent assay (ELISA) (Dia lab® HBs Ag, Austria) according to the manufacturer's instructions. Controls were defined by normal alanine aminotransferase (ALT) levels and hepatitis B core (HBc) antibody-positive/HbsAg-negative.
The inclusion criteria included those aged 15 - 100 years, being consented to take part in the study, possessing national identification documents, and having the ability to respond to the interviewer. Subjects who had hepatocellular carcinoma (HCC), HAV or HCV coinfection, hepatorenal syndrome, and alcohol history were excluded from our study. The ethics committee of the Faculty of Medicine, Birjand University, approved our work (ethics code ir.bums.REC.1395.55). Also, informed consent was obtained from all participants.
3.2. Genotyping
Two milliliters of blood specimens were gathered in tubes containing EDTA anticoagulant. Then, a salting-out method was used for the extraction of genomic DNA, and DNA purity and concentration were checked by a Nanodrop Spectrophotometer (Bio Tek, USA EPOCH). Extracted DNA was investigated for the -1082 A/G polymorphism by using polymerase chain reaction with restriction fragment length polymorphism (PCR-RFLP) (25). The primer sequences set were F: 5- TTCCCCAGGTAGAGCAACAC-3 and R: 5- TCC CTT ACT TTC CTC TTA CCT ATC C-3 (25).
Amplification was done in a volume of 25 µL containing 0.2 μL of 5 u/μL Taq DNA polymerase, 0.2 μL of 100 mM dNTP, 2.5 μL of 10X reaction buffer, 0.5 μL of 100 mM MgCl2, 1 μL of 100 pmoles/uL of each primer, 2 μL of template DNA, and 17.6 μL ddH2O. The PCR was performed in the Eppendorf PCR master cycler (AG-5345, Germany). After the initial denaturation at 95°C for 5 min, the second denaturation was done in 33 cycles of 95°C for 30 s, 58°C for 20 s, and 72°C for 25 s, with a final extension for 2 min at 72°C. For confirming successful amplification, 1.5% agarose gel was made in Tris/Borate/EDTA (TBE) buffer, and a 100-bp ladder was utilized for the presence of 171 base pair fragments. After that, digestion performed in 30-μL final volumes as follows: 18 μL nuclease-free water, 3 U restriction enzyme, 10 μL PCR product, and 2 μL 10 × Buffer. Tubes were incubated at 37°C for 10 h. The digestive fragments were observed on a 3% agarose gel stained with gel red.
3.3. Statistical Analysis
Statistical analysis was done using SPSS 16 software. The allele and genotype frequencies in two groups were calculated by direct counting, and the χ2 test with 95% Confidence Intervals (95% CI) from logistic regression analyses was utilized to acquire the Odds Ratio (OR) for chronicity of infection. Quantitative data are shown as mean ± standard deviation. A P value of <0.05 was considered to be significant.
4. Results
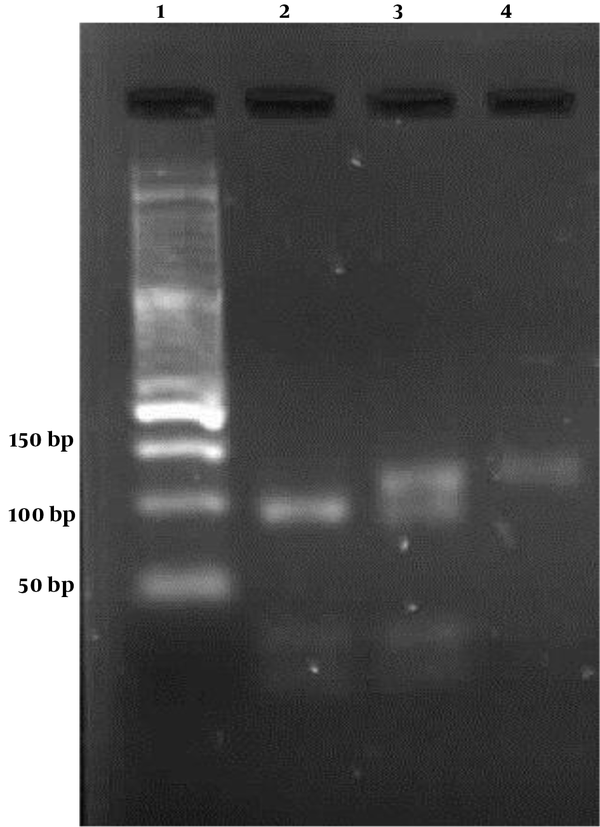
Our study enrolled 120 individuals, including 60 with CHB and 60 controls. The demographic characteristics of the control and case groups are compared in Table 1. There was no significant difference between the control and patient groups in age (P = 0.657) and gender (P = 0.855). The -1082 A/G polymorphism was recognized by enzyme digestion fragments (Figure 1). The IL-10 G allele gave four products of 21, 23, 32, and 95 base pairs, and IL-10 A allele gave three products of 118, 21, and 32 bp. The clearance of HBV infection demonstrated a significant association whit IL-10 -1082 polymorphisms just in the GG genotype. Reduced risk of chronic hepatitis B infection was associated with IL-10 -1082 GG (OR: 2.33, 95% CI: 1.07-5.09). However, IL-10 -1082 alleles did not have clear differences between the two study groups (Table 2).
Demographic Parameters of Two Study Groups
| Variable | Recovered Patients | CHB Patients | P Value |
|---|---|---|---|
| Age, y (mean ± SD) | 46.07 ± 15.68 | 47.25 ± 13.37 | 0.657 |
| Sex | 0.855 | ||
| Male | 32 | 34 | |
| Female | 28 | 26 |
Lane 1: 50 bp marker; lane 2: homozygous GG subjects (G allele digests with Mnl1 to produce 95 bp and 21, 23, 32 bp fragments); Lane 4: homozygous AA subjects (A allele digests with Mnl1 to produce 118 bp and 21 and 32 bp fragments); lane 3: heterozygous AG subjects
Genotype and Allele Frequency of Two Study Groups
| Genotype / Allele | CHB Patients (%) | Recovered Patients (%) | OR | 95 % CI | P Value |
|---|---|---|---|---|---|
| AA | 14 (23.3) | 12 (20) | 1 | ||
| AG | 37 (61.7) | 27 (45) | 0.73 | 0.44 - 1.19 | 0.21 |
| GG | 9(15) | 21 (35) | 2.33 | 1.07 - 5.09 | 0.03 |
| A | 65 (54.17) | 51 (42.5) | 1 | ||
| G | 55 (45.83) | 69 (57.5) | 1.59 | 0.96 - 2.66 | 0.07 |
5. Discussion
After HBV infection, the immune system induces to delete the virus. The majority of individuals become disease carriers, while some persons overcome the disease and become immune until the end of their lives. Part of carriers can become chronic hepatitis carriers and progress the disease to liver fibrosis and malignancy. The exact mechanisms clarifying these discrepancies have not been distinguished (14). But, several factors like viral load, genetic, age, sex, and immune status contribute to the variable clinical outcome of HBV infection (22, 26). The clearance of HBV is mediated by the secretion of pro-inflammatory and T helper1 cytokines and the prevention of virus replication. IL-10, whose gene polymorphisms can affect its expression, plays a principal function in the suppression of proinflammatory cytokine production (27, 28).
Recent studies indicated that -1082 GG, AG, and AA genotypes are related to high, intermediate, and low IL-10 production in HBV patients, respectively. In this study, we identified genotypes A/A, A/C, and C/C of IL-10-592 in the IL-10 promoter region and the correlation of this polymorphism with HBV susceptibility and recovery in the Birjand population. by comparison of chronic HBV infection subjects with spontaneously recovered controls, we found a significant difference only in IL-10-1082 GG between the two groups. This polymorphism showed a reduced risk of chronic HBV and it may have a role in the clearance of the virus. Furthermore, allele A was increased in CHB patients and allele G was increased in recovered HBV patients, but this difference was not significant. In contrast with our results, Gao et al. reported no significant difference in the frequency of the GG genotype while -1082 AA and AG genotypes were associated with an increased and reduced risk of persistent hepatitis B infection, respectively (29). But, like our study, there was no statistical significance in IL-10 -1082 A/G alleles between the two groups.
The study by Talaat et al. in an Egyptian population found that the GG genotype showed a significant increase in HBV patients than in healthy controls. Nevertheless, GA/AA genotypes were not significantly changed in the controls and HBV patients. In addition, the G allele could be accounted as a risk factor for HBV infection (30). Furthermore, a study performed in 2014 by Srivastva showed that the AA homozygous genotype was predominant in chronic cases and was associated with an increased risk of liver chronicity (18). But there are several studies in different populations throughout the world showing no difference in the -1082 position in the frequencies of alleles or genotype between the studied groups (13, 28, 31-33). It illustrates that patients and controls have a similar genetic background, and the IL-10 polymorphism does not influence the chronicity of HBV infection. By conducting a meta-analysis, Shu et al. demonstrated that the -1082 AA genotype was significantly associated with the self-clearance of hepatitis B following acute infection. In the subgroup analysis by ethnicity, they did not find a significant association between the -1082G/A variant and the spontaneous clearance of HBV infection in Asians and English-language studies (27).
The 1082G/A polymorphism has been reported to change gene expression. Turner et al. found that -1082 G was more related to the ‘high IL-10 producer’ phenotype than did the A allele. Also, the AA, AG, and GG genotypes had a relation to low, intermediate, and high IL-10 production in HBV patients, respectively (21). The equilibrium between the inflammatory and humoral response and immune regulation is determined by the levels of IL-10 (29). Low protein production has been reported to have a protective effect against HBV infection (34) because of a decrease in viral replication in chronic HBV infection (18).
Consistent with our results (the high frequency of GG genotype in the recovered group), Miyazoe et al. (35) reported that asymptomatic carriers are associated with low-producer genotype, and spontaneously recovered patients are associated with a high-producer genotype that has a lower HBV viral load and earlier HBeAg seroconversion (36), which disagrees with the view regarding the influence of interleukin 10 on the pathogenesis of HBV infection. These conflicting findings could be because of the impression of varied genes on HBV progression, geographical, or epidemiological factors. Moreover, they may depend on HBV genotype variations and also study circumstances like the characteristics of participants and sample size.
5.1. Conclusions
In summary, the polymorphisms of the IL-10-1082 GG genotype may be associated with the recovery of HBV in the Birjand population. In addition, IL-10 A/G alleles were not significantly different between chronic HBV patients and controls. However, because of the limited number of subjects in our study, a conclusion cannot be reached concerning the correlation of IL-10 gene promoter polymorphisms with HBV infection outcomes. Therefore, more investigations should be performed to determine this association.
References
-
1.
Locarnini S, Hatzakis A, Chen DS, Lok A. Strategies to control hepatitis B: Public policy, epidemiology, vaccine and drugs. J Hepatol. 2015;62(1 Suppl):S76-86. [PubMed ID: 25920093]. https://doi.org/10.1016/j.jhep.2015.01.018.
-
2.
Alavian SM, Hajarizadeh B, Ahmadzad-Asl M, Kabir A, Bagheri-Lankarani K. Hepatitis B virus infection in Iran: A systematic review. Hepat mont. 2008;8(4):281-94.
-
3.
WHO. Regional action plan for the implementation of the global health sector strategy on viral hepatitis 2017-2021. 2017. Available from: https://apps.who.int/iris/handle/10665/258729.
-
4.
Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095-128. https://doi.org/10.1016/s0140-6736(12)61728-0.
-
5.
Alter MJ. Epidemiology of viral hepatitis and HIV co-infection. J Hepatol. 2006;44(1 Suppl):S6-9. [PubMed ID: 16352363]. https://doi.org/10.1016/j.jhep.2005.11.004.
-
6.
Ren H, Zhang TT, Hu WL. A -819 C/T polymorphism in the interleukin-10 promoter is associated with persistent HBV infection, but -1082 A/G and -592A/C polymorphisms are not: a meta-analysis. Arch Virol. 2015;160(3):747-56. [PubMed ID: 25583543]. https://doi.org/10.1007/s00705-014-2317-7.
-
7.
Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30(12):2212-9. [PubMed ID: 22273662]. https://doi.org/10.1016/j.vaccine.2011.12.116.
-
8.
Wu JF, Wu TC, Chen CH, Ni YH, Chen HL, Hsu HY, et al. Serum levels of interleukin-10 and interleukin-12 predict early, spontaneous hepatitis B virus e antigen seroconversion. Gastroenterology. 2010;138(1):165-72 e1-3. [PubMed ID: 19782084]. https://doi.org/10.1053/j.gastro.2009.09.018.
-
9.
Duffy D, Mamdouh R, Laird M, Soneson C, Le Fouler L, El-Daly M, et al. The ABCs of viral hepatitis that define biomarker signatures of acute viral hepatitis. Hepatology. 2014;59(4):1273-82. [PubMed ID: 24500966]. https://doi.org/10.1002/hep.26901.
-
10.
Chen DQ, Zeng Y, Zhou J, Yang L, Jiang S, Huang JD, et al. Association of candidate susceptible loci with chronic infection with hepatitis B virus in a Chinese population. J Med Virol. 2010;82(3):371-8. [PubMed ID: 20087947]. https://doi.org/10.1002/jmv.21716.
-
11.
Thursz M, Yee L, Khakoo S. Understanding the host genetics of chronic hepatitis B and C. Semin Liver Dis. 2011;31(2):115-27. [PubMed ID: 21538279]. https://doi.org/10.1055/s-0031-1276642.
-
12.
Moudi B, Heidari Z, Mahmoudzadeh-Sagheb H, Hashemi M, Metanat M, Khosravi S, et al. Association between IL-10 gene promoter polymorphisms (-592 A/C, -819 T/C, -1082 A/G) and susceptibility to HBV infection in an Iranian population. Hepat Mon. 2016;16(2). e32427. [PubMed ID: 27148384]. [PubMed Central ID: PMC4852092]. https://doi.org/10.5812/hepatmon.32427.
-
13.
Sofian M, Kalantar E, Aghakhani A, Hosseini S, Banifazl M, Eslamifar A, et al. No correlation between interleukin-10 gene promoter polymorphisms and hepatitis B virus infection outcome. Hepat Mon. 2013;13(5). e8803. [PubMed ID: 23922563]. [PubMed Central ID: PMC3734898]. https://doi.org/10.5812/hepatmon.8803.
-
14.
Gao QJ, Xie JX, Wang LM, Zhou Q, Zhang SY. Interaction effects among IFN-gamma+874, IL-2-330, IL-10-1082, IL-10-592 and IL-4-589 polymorphisms on the clinical progression of subjects infected with hepatitis B virus and/or hepatitis C virus: a retrospective nested case-control study. BMJ Open. 2017;7(8). e013279. [PubMed ID: 28838891]. [PubMed Central ID: PMC5577879]. https://doi.org/10.1136/bmjopen-2016-013279.
-
15.
Lu YL, Wu X, Huang HL, Dai LC. Allele polymorphisms of interleukin-10 and hepatitis B, C virus infection. Chin Med J (Engl). 2010;123(10):1338-44. [PubMed ID: 20529592].
-
16.
Saxena R, Chawla YK, Verma I, Kaur J. Association of interleukin-10 with hepatitis B virus (HBV) mediated disease progression in Indian population. Indian J Med Res. 2014;139(5):737-45. [PubMed ID: 25027084]. [PubMed Central ID: PMC4140039].
-
17.
Westendorp RG, Langermans JA, Huizinga TW, Elouali AH, Verweij CL, Boomsma DI, et al. Genetic influence on cytokine production and fatal meningococcal disease. Lancet. 1997;349(9046):170-3. [PubMed ID: 9111542]. https://doi.org/10.1016/s0140-6736(96)06413-6.
-
18.
Srivastava M, Ranjan A, Choudhary JK, Tripathi MK, Verma S, Dixit VK, et al. Role of proinflammatory cytokines (interferon gamma) and anti-inflammatory cytokine (interleukin-10) gene polymorphisms in chronic hepatitis B infection: an Indian scenario. J Interferon Cytokine Res. 2014;34(7):547-51. [PubMed ID: 24446686]. [PubMed Central ID: PMC4080861]. https://doi.org/10.1089/jir.2013.0054.
-
19.
Hollegaard MV, Bidwell JL. Cytokine gene polymorphism in human disease: on-line databases, Supplement 3. Genes Immun. 2006;7(4):269-76. [PubMed ID: 16642032]. https://doi.org/10.1038/sj.gene.6364301.
-
20.
Eskdale J, Kube D, Tesch H, Gallagher G. Mapping of the human IL10 gene and further characterization of the 5' flanking sequence. Immunogenetics. 1997;46(2):120-8. [PubMed ID: 9162098]. https://doi.org/10.1007/s002510050250.
-
21.
Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet. 1997;24(1):1-8. [PubMed ID: 9043871]. https://doi.org/10.1111/j.1365-2370.1997.tb00001.x.
-
22.
Sodsai P, Surakiatchanukul T, Kupatawintu P, Tangkitvanich P, Hirankarn N. Association of cytokine and cytokine receptor gene polymorphisms with the risk of chronic hepatitis B. Asian Pac J Allergy Immunol. 2013;31(4):277-85. [PubMed ID: 24383970]. https://doi.org/10.12932/AP0284.31.4.2013.
-
23.
Ziaee M, Ebrahimzadeh A, Azarkar Z, Namaei MH, Saburi A, Fereidouni M, et al. Seroprevalence and risk factors for hepatitis B in an adult population: The first report from Birjand, South Khorasan, Iran. Hepat Mon. 2016;16(9). e36452. [PubMed ID: 27822260]. [PubMed Central ID: PMC5090805]. https://doi.org/10.5812/hepatmon.36452.
-
24.
Safari H, Anani Sarab G, Fereidouni M, Ziaee M, Mahavar N, Naghizadeh MS, et al. The CCR5-∆32 mutation: Impact on disease outcome in individuals with hepatitis B infection in the Southern Khorasan population (East of Iran). Hepat Mon. 2017;17(10). https://doi.org/10.5812/hepatmon.55014.
-
25.
Naranjo-Galvis CA, de-la-Torre A, Mantilla-Muriel LE, Beltran-Angarita L, Elcoroaristizabal-Martin X, McLeod R, et al. Genetic polymorphisms in cytokine genes in Colombian patients with ocular toxoplasmosis. Infect Immun. 2018;86(4). [PubMed ID: 29426041]. [PubMed Central ID: PMC5865049]. https://doi.org/10.1128/IAI.00597-17.
-
26.
Zhang TC, Zhang WF, Zhao YQ, Pan FM, Gao YF, Yuan H, et al. Gene variation in IL10 and susceptibility to chronic hepatitis B. J Infect. 2014;69(1):75-80. [PubMed ID: 24631780]. https://doi.org/10.1016/j.jinf.2014.03.003.
-
27.
Shu C, Wang J, He Y, Song T, Chen Z, Tang S, et al. Effects of interleukin 10 polymorphisms on the development of hepatitis B virus infection: a systemic review and meta-analysis. Int J Clin Exp Med. 2015;8(8):12028-40. [PubMed ID: 26550115]. [PubMed Central ID: PMC4612800].
-
28.
Gao L, Chen X, Zhang L, Wu D, Zhao H, Niu J. Association of IL-10 polymorphisms with hepatitis B virus infection and outcome in Han population. Eur J Med Res. 2016;21(1):23. [PubMed ID: 27245049]. [PubMed Central ID: PMC4888478]. https://doi.org/10.1186/s40001-016-0218-9.
-
29.
Gao QJ, Liu DW, Zhang SY, Jia M, Wang LM, Wu LH, et al. Polymorphisms of some cytokines and chronic hepatitis B and C virus infection. World J Gastroenterol. 2009;15(44):5610-9. [PubMed ID: 19938203]. [PubMed Central ID: PMC2785066]. https://doi.org/10.3748/wjg.15.5610.
-
30.
Talaat RM, Dondeti MF, El-Shenawy SZ, Khamiss OA. Association between IL-10 gene promoter polymorphism and hepatitis B viral infection in an Egyptian population. Biochem Genet. 2014;52(9-10):387-402. [PubMed ID: 24838671]. https://doi.org/10.1007/s10528-014-9655-8.
-
31.
Ribeiro CS, Visentainer JE, Moliterno RA. Association of cytokine genetic polymorphism with hepatitis B infection evolution in adult patients. Mem Inst Oswaldo Cruz. 2007;102(4):435-40. [PubMed ID: 17612762]. https://doi.org/10.1590/s0074-02762007005000043.
-
32.
Karatayli SC, Ulger ZE, Ergul AA, Keskin O, Karatayli E, Albayrak R, et al. Tumour necrosis factor-alpha, interleukin-10, interferon-gamma and vitamin D receptor gene polymorphisms in patients with chronic hepatitis delta. J Viral Hepat. 2014;21(4):297-304. [PubMed ID: 24597698]. https://doi.org/10.1111/jvh.12139.
-
33.
Yan Z, Tan W, Zhao W, Dan Y, Wang X, Mao Q, et al. Regulatory polymorphisms in the IL-10 gene promoter and HBV-related acute liver failure in the Chinese population. J Viral Hepat. 2009;16(11):775-83. [PubMed ID: 19413695]. https://doi.org/10.1111/j.1365-2893.2009.01139.x.
-
34.
Kallas E, Huik K, Pauskar M, Jogeda EL, Karki T, Des Jarlais D, et al. Influence of interleukin 10 polymorphisms -592 and -1082 to the HIV, HBV and HCV serostatus among intravenous drug users. Infect Genet Evol. 2015;30:175-80. [PubMed ID: 25542814]. https://doi.org/10.1016/j.meegid.2014.12.023.
-
35.
Miyazoe S, Hamasaki K, Nakata K, Kajiya Y, Kitajima K, Nakao K, et al. Influence of interleukin-10 gene promoter polymorphisms on disease progression in patients chronically infected with hepatitis B virus. Am J Gastroenterol. 2002;97(8):2086-92. [PubMed ID: 12190181]. https://doi.org/10.1111/j.1572-0241.2002.05926.x.
-
36.
Wu JF, Ni YH, Lin YT, Lee TJ, Hsu SH, Chen HL, et al. Human interleukin-10 genotypes are associated with different precore/core gene mutation patterns in children with chronic hepatitis B virus infection. J Pediatr. 2011;158(5):808-13. [PubMed ID: 21168854]. https://doi.org/10.1016/j.jpeds.2010.11.015.
