Abstract
Background:
Epithelial ovarian cancer (EOC) is a deadly gynecological cancer among women worldwide. Ministering ovarian cancer with classic chemotherapy medications can cause various side effects. Thus, in recent years, the anticancer potential of medicinal plants has been noticed in complementary cancer treatment due to their high efficacy and low toxicity. The anti-proliferative potential of Xanthium strumarium leaf and seed extract against several cancer cell lines was reported; however, there is no report on the effect of its root so far.Objectives:
In this study, the antitumor effect of X. strumarium root extract was evaluated on the SK-OV-3 ovarian cancer cell line, and metabolic alterations were identified.Methods:
Cancer cells were cultured and ministered with different concentrations of ethanolic root extract. The antitumor effect of the root extract was determined by MTT assay, and cell metabolites was extracted. Finally, the metabolome profile was characterized and analyzed by 1H NMR-based metabolomics.Results:
The IC50 value of the root extract of X. Strumarium was 30 µg/mL after 48 hours, which exhibited anticancer activity against SK-OV-3 ovarian cancer cells. Aminoacyl-tRNA synthesis, glycerolipid metabolism, fatty acid biosynthesis, and biotin metabolism were the most affected metabolic pathways involved in the growth inhibition of cancer cells.Conclusions:
In the current study, the ethanolic root extract of X. Strumarium reveals antitumor activity against SK-OV-3 ovarian cancer cells and could affect vital metabolic pathways.Keywords
Xanthium strumarium Epithelial Ovarian Cancer SK-OV-3 Cell Line Metabolomics 1H Nuclear Magnetic Resonance
1. Background
Epithelial ovarian cancer (EOC) is one of the women’s most typical and highly malignant gynecological cancers. Globally, the number of women with ovarian cancer is about 313959, with 207252 deaths yearly (1). The American Cancer Society, in 2021, declared that about 109000 women were diagnosed with ovarian cancer in the USA, among which about 33000 patients had expired (2). Around 90% of all ovary, fallopian tubes, and primary peritoneal cancers are epithelial tumors. There are three forms of standard therapy for advanced ovarian cancer: Surgery, chemotherapy, and radiation. The considerably effective chemotherapy drugs to treat ovarian cancer are platinum analogs such as cisplatin and carboplatin after the optimal surgical cytoreduction (3). However, the disease returns in most patients with progressive ovarian cancer due to a boost in serum CA125 levels (4). Many reports have stated that ministering ovarian cancer with established chemotherapy drugs can induce various side effects. In contrast, natural compound-based drugs can affect invasive cancer growth with fewer side effects. Therefore, researchers have recently concentrated on the potential use of medicinal plants as biological anticancer agents due to their high biological activity and low toxicity (5).
Xanthium strumarium is a medicinal plant naturally found as a weed that belongs to the Asteraceae (Compositae) family. It has been extensively naturalized in Brazil, Malaysia, China, India, North America, and Iran. Many investigations have reported the various pharmacological activity of the X. strumarium, such as anticancer effects (6, 7). The anti-proliferative potential of X. strumarium leaf extract against several cancer cell lines was reported, i.e., SK-MEL-2 (Melanoma), A549 (lung), SK-OV-3 (ovary), HTC-15 (colon) and XF498 (CNS) (8, 9). It has been found that the seed extract of X. strumarium could also inhibit the growth of the L929, HEPG2, A549, and Jurkat cell lines (10). The anticancer effect of X. Strumarium root extract on S180 and HEPG2 cancer cell lines has likewise been established (10). Coumarins, a phenylpropanoid, were found just in the roots of this plant rather than in other parts of this plant. It has been revealed that coumarin derivatives could inhibit the growth of OVCAR-3(ovary), HL-60 (leukemia), MCF7 (breast), H727(lung), A549 (lung), ACHN (renal), and many cancer cell lines (11). Coumarins also could counteract the side effects caused by radiotherapy (12). As the root of X. strumarium is enriched with coumarin, our experiment evaluated the anti-proliferation potential of X. strumarium root extract against an ovarian cancer cell line.
Metabolomics is an analytical profiling approach for estimating and comparing vast numbers of metabolites in biological samples (13). NMR-based metabolomics is an emerging technology utilized to describe cancer metabolism, with potential use in clinical diagnosis and follow-up of cancer treatment (14).
2. Objectives
The current investigation focuses on the anticancer activity of root extract of X. strumarium on the human ovarian cancer cell line Skov3 to see if it could successfully influence the growth of these epithelial ovarian cancer cells. Eventually, the metabolome profile and the most significant metabolic pathways influenced by root extract are examined to determine how this extract might affect the disease. However, this data output will be used for further investigation in drug modeling in combination with other omics data.
3. Methods
3.1. Plant Extract
Xanthium strumarium roots were gathered from the Kermanshah region, Iran, and authenticated by the Central Herbarium of Tehran University under voucher No 48241. Roots were rinsed, dried, and grounded. Roots were extracted with 500 mL of 80% ethanol by the Soxhlet apparatus. The extract was cleaned with charcoal to remove the color pigments. A Rotary evaporator removed the solvent, and the aliquots of pure extract were stored in a sterile receptacle at 4°C for further use.
3.2. Cancer Cell Culture
SK-OV-3 (human ovarian cancer cell line) was received from the Pasteur Institute of Iran. Cancer cells were cultured in RPMI-1640 complete medium supplemented with 10% FBS and penicillin/streptomycin (100X) as an antibiotic. Cells were maintained and grown in a CO2 incubator at 37°C until they reached > 85% confluence
3.3. Measurement of Total Phenolic Content
The total phenolic content (TPC) of X. Strumarium root extract was determined using the Folin-Ciocalteu assay, and Gallic acid was used as the external calibration standard (15).
3.4. MTT Assay
The impact of root extract on cell viability was estimated using 3-(4,5 dimethylthiazol-2-yl)-2,5 diphenyl tetrazolium bromide (MTT) assay. Eight thousand cells/well were seeded and incubated for 24 hour at 37°C. The root extract was added (600, 60, 30, 20, 15, 12, and 6 µg/mL) in each well. After 48 hours, 20 µL MTT solution was added to each well and incubated for 3 hours. Also, 200 µL of DMSO was added and shaken for 20 min to dissolve the formazan crystals. The absorbance was taken at 540 nm by Eliza Reader. Three independent experiments assessed all the tests and control (PBS) in triplicate. The half-maximal inhibitory concentration (IC50) was calculated using the following formula:
3.5. Cell Extraction
Five control and 5 test samples were prepared, containing 70 × 106 cells/mL each. According to the associated IC50 value, test samples were treated with 9000 µg/mL of root extract, incubated, trypsinized, and centrifuged at 1000 rpm (4°C) for 5 min. To each sample, PBS was added and centrifuged again. Then 0.5 mL of methanol and 1 mL of cold chloroform were added to every 107 cells, and samples were vortexed. An equal volume of chloroform: Deionized distilled water was added to the samples, sonicated (10 minutes), and centrifuged at 4000 rpm (4°C) for 15 min, leading to the formation of two layers. The top hydrophilic and bottom lipophilic layers were separated, transferred to clean tubes, and lyophilized to achieve the 1H NMR spectrum (16).
3.6. 1H NMR Spectroscopy
Samples were resuspended in 700 µL of 100mM phosphate buffer (pH 7.0) prepared in D2O, having one mM TSP as an internal reference and 2 mM imidazole as a pH indicator. Lipophilic samples were resuspended in 700 µL of deuterated chloroform. Samples were centrifuged at 10000 rpm for 10 minutes at 4°C. 500 µL of the extracted samples were moved to NMR probes and scrutinized on a Bruker AV-500 NMR spectrometer with a field gradient operating at 500.13 MHz for proton observation at 298 K. One-dimensional 1H NMR spectra were registered using a 10- µs pulse, 0.1 s mixing time, 3.0 s relaxation delay, 6009.6 Hz spectral width, and 3000 transients with standard 1D NOESY pulse sequence to suppress the residual water peak (17).
3.7. Data Analysis
Spectra were preprocessed utilizing the ProMetab function file (V.3.3) in the MATLAB package (v.7.8.0.347). In the following, each spectrum was aligned and binned at 0.005 ppm. The peak at 4.7 ppm (water) was eliminated. Multivariate data analysis was conducted using the classification PLS-DA method. The discrimination chemical shifts were reported as score plots, loading plots, and VIP scores. Projected chemical shifts were taken to the HMDB database to find outliers, and corresponding metabolites were specified. MetaboAnalyst (V.4.0), a set of online tools for metabolomics data analysis and interpretation, was employed to specify the altered metabolic pathways in this investigation.
4. Results
4.1. Total Polyphenol Content
The total phenolic content of the root extract of X. Strumarium was 6000 µg/mL.
4.2. Proliferation Assay
The IC50 value of the root extract of X. Strumarium was 31.7 µg/mL (Table 1), which showed anticancer activity against SK-OV-3 ovarian cancer cells.
| Extract Concentration | % Cell Viability Test 1 | % Cell Viability Test 2 | % Cell Viability Test 3 |
|---|---|---|---|
| 0 µg/mL + PBS (standard) | 100 | 100 | 100 |
| 6 | 97 | 100 | 96 |
| 12 | 81 | 91 | 81 |
| 15 | 75 | 82 | 73 |
| 20 | 63 | 64 | 63 |
| 30 | 48 | 46 | 48 |
| 60 | 37 | 40 | 39 |
| 600 | 13 | 20 | 32 |
| 31.7 | 50 | - | - |
4.3. Effect of Xanthium strumarium Root Extract on Metabolome Profile
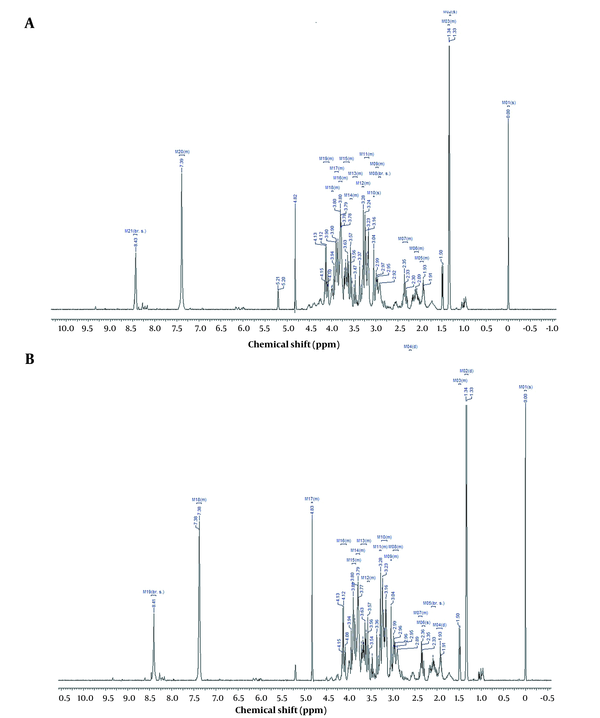
1H NMR spectroscopy exhibited that the root extract of X. Strumarium influenced metabolites (Tables 2 and 3) and metabolic pathways (Figure 1). PLS-DA analysis leads to data classification in this research, displayed as loading plots among the selected components (Figure 2).
Metabolites Altered in Hydrophilic Group
| Metabolites | HMDB (No.) |
|---|---|
| 1. Hypo taurine | HMDB0000965 |
| 2. Tetrahydrofolic acid | HMDB0001846 |
| 3. 5-(2-Hydroxyethyl)-4-methyl-thiazole | HMDB0032985 |
| 4. Glycerol | HMDB0000131 |
| 5. O-tyrosine | HMDB0006050 |
| 6. 2-Amino-3-phosphonopropionic acid | HMDB0000370 |
| 7. Deoxyadenosine | HMDB0000101 |
| 8. Coenzyme A | HMDB0001423 |
| 9. Rhamnose | HMDB0000849 |
| 10. Erythrose | HMDB0002649 |
| 11. Beta-leucine | HMDB0003640 |
| 12. Gluconic acid | HMDB0000625 |
| 13. D-xylitol | HMDB0002917 |
| 14. Cysteine glutathione disulfide | HMDB0000656 |
| 15. L-tyrosine | HMDB0000158 |
| 16. Ethen deoxyadenosine | HMDB0001786 |
| 17. D-alanine | HMDB0001310 |
| 18. L-alanine | HMDB0000161 |
| 19. Tryptamine | HMDB0000303 |
| 20. 14-beta-D-glucan | HMDB0006944 |
| 21. Methylmalonic acid | HMDB0000202 |
| 22. 15-anhydrosorbitol | HMDB0002712 |
| 23. D-arabitol | HMDB0000568 |
| 24. A-Ketoglutaric acid oxime | HMDB0002467 |
| 27. Methyl isothiocyanate | HMDB0034106 |
| 28. Glycerol 3-phosphate | HMDB0000126 |
| 30 D-ribonolactone | HMDB0001900 |
| 31. Gamma-cyclodextrin | HMDB0029927 |
| 32. 5-Thymidylic acid | HMDB0001227 |
| 33. 4-hydroxyproline | HMDB0000725 |
| 34. D-mannose | HMDB0000169 |
| 35. L (-)-nicotine | HMDB0001934 |
| 36. L-proline | HMDB0000162 |
| 37. D-galactose | HMDB0000143 |
| 38. Chlormezanone | HMDB0015309 |
| 39. 5-methoxytryptophol | HMDB0001896 |
| 40. Hydroxypropionic acid | HMDB0000700 |
| 41. L-homoserine | HMDB0000719 |
| 43. 2 3-Dihydroxyvaleric acid | HMDB0000421 |
| 44. L-glutamine | HMDB0000641 |
| 45. 5-hydroxytryptophol | HMDB0001855 |
| 46. Ribitol | HMDB0000508 |
| 47. Propylene glycol | HMDB0001881 |
Metabolites Altered in Lipophilic Group
| Metabolites | HMDB (No.) |
|---|---|
| 1. Caproic acid | HMDB0000535 |
| 2. 2-oxohexane | HMDB0005842 |
| 3. Propane | HMDB0031630 |
| 4. Valeric acid | HMDB0000892 |
| 5. 3-methylheptane | HMDB0031583 |
| 6. Rhamnose | HMDB0000849 |
| 7. Decanal 1 | HMDB0011623 |
| 8. 2-heptanone | HMDB0003671 |
| 9. Hexacosanoic acid | HMDB0002356 |
| 10. Heptanoic acid | HMDB0000666 |
| 11. Heneicosanoic acid | HMDB0002345 |
| 12. Diethylthiophosphate | HMDB0001460 |
| 13. Octanol | HMDB0001183 |
| 14. Heptadecanoic acid | H MDB0002259 |
| 15. Nonivamide | HMDB0029846 |
| 16. Octadecanol | HMDB0002350 |
| 17. Elaidic acid | HMDB0000573 |
| 18. Pantothenic acid | HMDB0000210 |
| 19. Lignocerane | HMDB0034282 |
| 20. Nonacosane | HMDB0034288 |
| 21. Nonadecane | HMDB0034289 |
| 22. Tricosanoic acid | HMDB0001160 |
| 23. Arachidic acid | HMDB0002212 |
| 24. Pimelic acid | HMDB0000857 |
| 27. Ethyl tetradecanoate | HMDB0034153 |
| 28. 5-methoxydimethyltryptamine | HMDB0002004 |
| 30. 1-octacosanol | HMDB0034380 |
| 31. Hexadecane | HMDB0033792 |
| 32. Nonadecanoic acid | HMDB0000772 |
| 33. Cis-aconitic acid 1 | HMDB0000072 |
| 34. Capric acid | HMDB0000511 |
| 35. 2-dodecanone 1 | HMDB0031019 |
| 36. Pelargonic acid | HMDB0000847 |
| 37. L-tyrosine 1 | HMDB0000158 |
| 38. Docosanol 1 | HMDB0014770 |
| 39. Tridecane 1 | HMDB0034284 |
| 40. Undecane 1 | HMDB0031445 |
| 41. Oenanthic ether | HMDB0000798 |
| 42. 17-epiestriol 1 | HMDB0000356 |
| 43. Valproic acid 1 | HMDB0001877 |
| 44. Octacosanoic acid | HMDB0002348 |
| 45. Glycogen | HMDB0000757 |
| 46. L-isoleucine | HMDB0000172 |
| 47. 1-pentadecene | HMDB0031082 |
| 48. Stearic acid | HMDB0000827 |
| 49. Naringin | HMDB0002927 |
| 50. Dl-2-aminooctanoic acid 1 | HMDB0000991 |
| 51. Oleic acid | HMDB000207 |
4.4. Pathway Analysis
HMDB database was used to find the significant metabolites associated with the NMR chemical shifts. Also, affected metabolic pathways related to these separated outliers were identified by the MetaboAnalyst online tools and the KEGG database. Tables 4 and 5 exhibit metabolome analysis results. The most important pathway was taken for the discussion section.
Pathway Analysis for Hydrophilic Groups of SK-OV-3 Ovarian Cancer Cells Were Treated by Xanthium strumarium Root Extract a
| Pathways, Metabolites | Total | Hits | Raw P |
|---|---|---|---|
| Aminoacyl-tRNA synthesis | 75 | 5 | 0.0088 |
| L-glutamine | |||
| L-alanine | |||
| L- tyrosine | |||
| L-proline | |||
| Tetrahydro folic acid | |||
| Glycerolipid metabolism | |||
| Glycerol 3-phosphate glycerol Propylene glycol | 32 | 3 | 0.0172 |
| Taurine and hypotaurine metabolism | |||
| Hypotaurine L-alanine | 20 | 2 | 0.0463 |
| Pentose and glucoronate interconversion | |||
| Ribitol D-xylitol D-arabitol | 53 | 3 | 0.0633 |
| Thiaminmetabolism | |||
| L-tyrosine 5-(2hydroxyl)-4 methyl thiazol | 24 | 2 | 0.0644 |
| Alain aspartate and glutamate metabolism | |||
| L-glutamine L-alanine | 24 | 2 | 0.0644 |
| Pyrimidine metabolism | |||
| L-glutamine methylmalonic acid 5-thymidylic acid | 60 | 3 | 0.0851 |
Pathway Analysis for Lipophilic Groups of SK-OV-3 Ovarian Cancer Cells Were Treated by Xanthium strumarium Root Extract a
| Pathways, Metabolites | Total | Hit | Raw P |
|---|---|---|---|
| Fatty acid biosynthesis | |||
| Capric acid, oleic acid, stearic acid | 49 | 3 | 0.0238 |
| Biotin metabolism | |||
| Pimelic acid | 11 | 1 | 0.1331 |
| Citrate cycle (TCA cycle) | |||
| Cis-aconitic acid | 20 | 1 | 0.2291 |
| Aminoacyl-tRNA synthesis | |||
| Leucine, L-tyrosine | 75 | 2 | 0.2513 |
| Thiamin metabolism | |||
| L- tyrosine | 24 | 1 | 0.2684 |
1H NMR spectra of hydrophilic (A) and lipophilic (B) of SK-OV-3 ovarian cancer cells
PLS-DA loading plot for hydrophilic (A) and lipophilic (B) groups of SK-OV-3 ovarian cancer cells

The loading 1-axis shows the magnitude of the spectral bins. The loading 2-axis indicates the correlation of the bins to the predictive variation. The collected spots were the shared metabolites in both cell groups, while scattered spots were targeted metabolites in the current study.
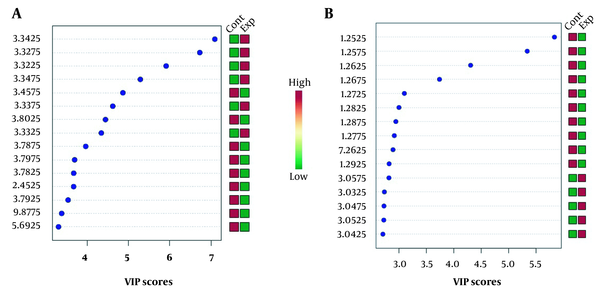
VIP score measures the variable’s importance (chemical shifts) in the PLS-DA model (Figure 3). The Y-axis displays the VIP scores associated with each variable on the X-axis. The colored boxes on the right demonstrate the metabolite concentrations in the experimental and control cell groups.
VIP scores for hydrophilic (A) and lipophilic (B) groups of SK-OV3 ovarian cancer cells
5. Discussion
Cancer cells require several vital nutrients to reach their changing metabolic needs as they progress through stages of development compared to normal cells. Our findings depict that the root extract of X. Strumarium has favorable anticancer activity in the epithelial ovarian cancer SK-OV-3 cell line. Like many other cancer types, epithelial ovarian cancer reprograms its metabolism during progression, converting from oxidative phosphorylation to glycolysis, a phenomenon recognized as the Warburg effect (17). Alterations in several metabolic pathways, such as glycolysis, the tricarboxylic acid cycle, amino-acid metabolism, and lipid metabolism, were seen in the sera of epithelial ovarian cancer patients. These metabolic pathways are bonded, so their disruption leads to the initiation and progression of cancer (18).
In this investigation, various metabolites altered after the cells were treated with root extract of X. Strumarium. Plotting these altered metabolites showed that Aminoacyl-tRNA synthesis, glycerolipid metabolism, fatty acid biosynthesis, and Biotin metabolism were the most affected metabolic pathways in cell growth inhibition.
The absorption of amino acids in cancer cells is faster than in healthy cells. It has been declared that balanced amino acid nutrients support the proliferation of cancer cells, whereas unbalanced amino acid nutrients suppress tumor cell proliferation (19). It was reported that the reduction of essential amino acids leads to a disturbance in the metabolism of cancer cells and prevents the growth of tumors by inhibiting protein synthases (20). The attachment of amino acids to specific tRNAs in protein synthesis is catalyzed by aminoacyl tRNA synthases, which play an essential role in the survival of cancer cells. Aminoacyl tRNA is the first protein biosynthesis product (21). It was exposed that the destruction of leucyl tRNA synthases in lung cancer cells reduced the ability of cell migration and colony formation (22). It has also been reported that increased expression of leucyl tRNA synthases was found in cancer cells, which seems to be recognized as a potential metabolic target (23). Leucyl tRNA synthase is associated with the AIMP1 protein and other multi-functional proteins. In genotoxicity, these proteins stimulate the P53 activity, then adjust the P21 expression. P21 protein is an essential intermediate through which P53 promotes cell cycle inhibition (24). It has been stated that serum tryptophanyl levels are related to colon cancer cells’ survival (25). For glioblastoma multiforme tumors, it has been shown that the expression of several ARS genes, including CysRS, ASnRs, PhenRS, GlnRS, and ValRS, leads to more growth of cancer cells (26). Also, it was proposed that increased expression of the ThrRS gene is directly related to the risk of ovarian, breast, and lung cancers (27). In animal models, it was found that high levels of MetRS can be a marker for an increased risk of death due to lung cancer (28). It was also reported that anti-ThRS could inhibit the growth of Jurkat and CEM cancer cell lines compared to control cells (26). In this research, the metabolites of L-glutamine, L-alanine, L-tyrosine, L-proline, and Tetrahydrofolic acid altered the aminoacyl tRNA biosynthesis pathway. A comparison of our results with previous findings indicated that the root extract of X. strumarium could disrupt the activity of tRNA synthase enzymes, thus, restricting the formation of the aminoacyl tRNAs, which inhibits the growth of SK-OV-3 ovarian cancer cells. Leucyl tRNA synthase and glutamyl tRNA synthase have more potential for metabolic anticancer targets than other enzymes.
Tetrahydrofolic acid, one of the metabolites altered in this pathway, is made by dihydrofolate reductase from folic acid, an inactive biochemical substance. Tetrahydrofolic acid enters the cells, used in red blood cell production, purine nucleic acid synthesis, thymidylate synthase metabolism, and amino acid metabolism (29). Therefore, it seems that X. strumarium root extract has inhibited the growth of the SK-OV-3 ovarian cancer cells, maybe by changing the tetrahydrofolic acid cofactor, the active form of folic acid, and also by disrupting the metabolism of amino acids.
Alterations in the normal metabolism of lipids, especially serum triglycerides, have been detected in patients with breast and ovarian cancer, which indicates a positive correlation between the level of triglycerides and the risk of progression of these cancers (30). A positive relationship was reported between serum triglycerides and esophageal, colorectal, lung, renal, and thyroid cancer (31). A partial connection was discovered between serum triglyceride levels and thyroid cancer in men and endometrial, cervical, and bladder cancer in women (32). Also, a direct correlation was found between serum triglyceride levels and gynecological cancers (33). In this research, Glycerol 3-phosphate, glycerol, and propylene glycol metabolites have changed in the glycerolipid pathway. According to the KEGG database, altered propylene glycol metabolite can motivate alcohol dehydrogenase enzyme, reducing propylene glycol and inhibiting the SK-OV-3 ovarian cancer cell line. Also, altered glycerol and glycerol 3-phosphate metabolites can cause disorders in fatty acid metabolism and the glycolysis cycle. Hence, it seems that X. strumarium root extract was able to inhibit the growth of SK-OV-3 ovarian cancer cells, probably by reducing their energy.
Variations in lipid metabolism were discovered in patients with ovarian and recurrent cancer (34). Fatty acid (FA) synthesis is the creation of fatty acids from acetyl-CoA and NADPH through the action of enzymes called fatty acid synthase (FASN). In cancer cells, most FAs are synthesized by FASN for essential changes. Therefore, FASN has been considered a marker for diagnosis, prognosis, and a therapeutic target in cancer (35). Palmitic acid promotes cell proliferation because of amplified expression of FASN in ovarian cancer cells, and suppression of this enzyme causes apoptosis and cancer cell death (36). It was reported that increased FASN gene expression occurs in many tumors such as breast, prostate, colon, and ovary due to the non-reaction of tumors to regulatory messages and their greater affinity towards the lipogenesis pathway (37). It has been revealed that stearic acid has a negative role in DNA damage that stimulates cell alteration and tumor genesis (36). It has also been detected that decreased levels of stearic acid and oleic acid in serum-initiated malignancy in cells (38).
It was reported that an increase in oleic acid and a decrease in stearic acid was shown in several cancer types such as breast, liver, lung, pancreas, colon, and prostate (39). It has also been exposed that oleic acid has inhibited the activity of FASN, and the malonyl CoA has accumulated, suppressing HER2 oncogene expression (40, 41).
This research alters oleic acid, capric acid, and stearic acid metabolites in fatty acid biosynthesis. Therefore, oleic and stearic acid can be potential therapeutic targets in ovarian cancer. A comparison of our results with previous findings revealed that X. Strumarium root extract could probably prevent the growth of SK-OV-3 cancer cells by inhibiting FASN activity.
Biotin is an essential cofactor for carboxylases made from pimelic acid, coenzyme A, ATP, and mg2+. It has been demonstrated that altered pimelic acid inhibits biotin production. Biotin disturbs the activity of the pyruvate carboxylase enzyme. It disrupts the gluconeogenesis pathway in cancer cells, leading to the inability of cancer cells to produce glucose and a decrease in cellular energy (42). It was also reported that altered pimelic acid had inhibited biotin synthesis, and the acetyl-CoA carboxylase enzyme has been disrupted, preventing malonyl CoA production, a substrate for fatty acid synthesis. Finally, the fatty acid biosynthesis pathway has been disturbed, and cancer cells’ energy has been reduced (43). In this research, pimelic acid metabolite has changed in the biotin metabolism pathway, so pimelic acid can be a potential therapeutic target in ovarian cancer. A comparison of our findings with previous results represented that X. strumarium root extract could probably inhibit the growth of SK-OV-3 ovarian cancer cells by disrupting the biotin metabolism pathway, disrupting fatty acid synthesis, and reducing cellular energy.
5.1. Conclusions
In the present investigation, the ethanolic root extract of X. Strumarium reveals antitumor activity against SK-OV-3 ovarian cancer cells. Lipid metabolism was the most affected metabolic pathway involved in cell growth inhibition and could be a potential medicine target in ovarian cancer treatment. However, further studies are required to validate these findings along with the various potential fractions of the root extract of this plant, in addition to the SK-OV-3 cell’s transcriptome and genomic data.
References
-
1.
Nag S, Aggarwal S, Rauthan A, Warrier N. Maintenance therapy for newly diagnosed epithelial ovarian cancer- a review. J Ovarian Res. 2022;15(1):88. [PubMed ID: 35902911]. [PubMed Central ID: PMC9331490]. https://doi.org/10.1186/s13048-022-01020-1.
-
2.
Islami F, Guerra CE, Minihan A, Yabroff KR, Fedewa SA, Sloan K, et al. American Cancer Society's report on the status of cancer disparities in the United States, 2021. CA Cancer J Clin. 2022;72(2):112-43. [PubMed ID: 34878180]. https://doi.org/10.3322/caac.21703.
-
3.
Schoutrop E, Moyano-Galceran L, Lheureux S, Mattsson J, Lehti K, Dahlstrand H, et al. Molecular, cellular and systemic aspects of epithelial ovarian cancer and its tumor microenvironment. Semin Cancer Biol. 2022;86(Pt 3):207-23. [PubMed ID: 35395389]. https://doi.org/10.1016/j.semcancer.2022.03.027.
-
4.
Charkhchi P, Cybulski C, Gronwald J, Wong FO, Narod SA, Akbari MR. CA125 and ovarian cancer: A comprehensive review. Cancers (Basel). 2020;12(12). [PubMed ID: 33322519]. [PubMed Central ID: PMC7763876]. https://doi.org/10.3390/cancers12123730.
-
5.
Ng JY, Lau SKC. Complementary and alternative medicine status in ovarian cancer guidelines: A systematic review. Eur J Integr Med. 2020;40:101227. https://doi.org/10.1016/j.eujim.2020.101227.
-
6.
Malekzadeh R, Arjmand M, Haji Hosseini R, Vaziri A, Zamani Z. Evaluation of the anticancer effect of xanthium strumarium root extract on human epithelial ovarian cancer cells using 1H NMR-based metabolomics. Journal of Sciences, Islamic Republic of Iran. 2020;31(3):205-12.
-
7.
Chang HW, Liu PF, Tsai WL, Hu WH, Hu YC, Yang HC, et al. Xanthium strumarium fruit extract inhibits ATG4B and diminishes the proliferation and metastatic characteristics of colorectal cancer cells. Toxins (Basel). 2019;11(6). [PubMed ID: 31159487]. [PubMed Central ID: PMC6628400]. https://doi.org/10.3390/toxins11060313.
-
8.
Francisco Fernandez M, Charfi C, Piloto-Ferrer J, Lidia Gonzalez M, Lamy S, Annabi B. Targeting ovarian cancer cell cytotoxic drug resistance phenotype with xanthium strumarium l. Extract. Evid Based Complement Alternat Med. 2019;2019:6073019. [PubMed ID: 31827554]. [PubMed Central ID: PMC6885198]. https://doi.org/10.1155/2019/6073019.
-
9.
Fan W, Fan L, Peng C, Zhang Q, Wang L, Li L, et al. Traditional uses, botany, phytochemistry, pharmacology, pharmacokinetics and toxicology of xanthium strumarium l.: A review. Molecules. 2019;24(2). [PubMed ID: 30669496]. [PubMed Central ID: PMC6359306]. https://doi.org/10.3390/molecules24020359.
-
10.
Al-Mekhlafi FA, Abutaha N, Mashaly AMA, Nasr FA, Ibrahim KE, Wadaan MA. Biological activity of xanthium strumarium seed extracts on different cancer cell lines and aedes caspius, culex pipiens (Diptera: Culicidae). Saudi J Biol Sci. 2017;24(4):817-21. [PubMed ID: 28490952]. [PubMed Central ID: PMC5415140]. https://doi.org/10.1016/j.sjbs.2016.07.003.
-
11.
Bhattarai N, Kumbhar AA, Pokharel YR, Yadav PN. Anticancer potential of coumarin and its derivatives. Mini Rev Med Chem. 2021;21(19):2996-3029. [PubMed ID: 33820507]. https://doi.org/10.2174/1389557521666210405160323.
-
12.
Schmidt DR, Patel R, Kirsch DG, Lewis CA, Vander Heiden MG, Locasale JW. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J Clin. 2021;71(4):333-58. [PubMed ID: 33982817]. [PubMed Central ID: PMC8298088]. https://doi.org/10.3322/caac.21670.
-
13.
Raja G, Jung Y, Jung SH, Kim T. 1H-NMR-based metabolomics for cancer targeting and metabolic engineering–A review. Process Biochem. 2020;99:112-22.
-
14.
Fernandes FH, Salgado HR. Gallic acid: Review of the methods of determination and quantification. Crit Rev Anal Chem. 2016;46(3):257-65. [PubMed ID: 26440222]. https://doi.org/10.1080/10408347.2015.1095064.
-
15.
Saborano R, Eraslan Z, Roberts J, Khanim FL, Lalor PF, Reed MAC, et al. A framework for tracer-based metabolism in mammalian cells by NMR. Sci Rep. 2019;9(1):2520. [PubMed ID: 30792403]. [PubMed Central ID: PMC6385278]. https://doi.org/10.1038/s41598-018-37525-3.
-
16.
Emwas AH, Roy R, McKay RT, Tenori L, Saccenti E, Gowda GAN, et al. NMR spectroscopy for metabolomics research. Metabolites. 2019;9(7). [PubMed ID: 31252628]. [PubMed Central ID: PMC6680826]. https://doi.org/10.3390/metabo9070123.
-
17.
Warburg O. On the origin of cancer cells. Science. 1956;123(3191):309-14. [PubMed ID: 13298683]. https://doi.org/10.1126/science.123.3191.309.
-
18.
Sreedhar A, Zhao Y. Dysregulated metabolic enzymes and metabolic reprogramming in cancer cells. Biomed Rep. 2018;8(1):3-10. [PubMed ID: 29399334]. [PubMed Central ID: PMC5772474]. https://doi.org/10.3892/br.2017.1022.
-
19.
Wei Z, Liu X, Cheng C, Yu W, Yi P. Metabolism of amino acids in cancer. Front Cell Dev Biol. 2020;8:603837. [PubMed ID: 33511116]. [PubMed Central ID: PMC7835483]. https://doi.org/10.3389/fcell.2020.603837.
-
20.
Sivanand S, Vander Heiden MG. Emerging roles for branched-chain amino acid metabolism in cancer. Cancer Cell. 2020;37(2):147-56. [PubMed ID: 32049045]. [PubMed Central ID: PMC7082774]. https://doi.org/10.1016/j.ccell.2019.12.011.
-
21.
Hyeon DY, Kim JH, Ahn TJ, Cho Y, Hwang D, Kim S. Evolution of the multi-tRNA synthetase complex and its role in cancer. J Biol Chem. 2019;294(14):5340-51. [PubMed ID: 30782841]. [PubMed Central ID: PMC6462501]. https://doi.org/10.1074/jbc.REV118.002958.
-
22.
Bian M, Huang S, Yu D, Zhou Z. tRNA metabolism and lung cancer: Beyond translation. Front Mol Biosci. 2021;8:659388. [PubMed ID: 34660690]. [PubMed Central ID: PMC8516113]. https://doi.org/10.3389/fmolb.2021.659388.
-
23.
Nie A, Sun B, Fu Z, Yu D. Roles of aminoacyl-tRNA synthetases in immune regulation and immune diseases. Cell Death Dis. 2019;10(12):901. [PubMed ID: 31780718]. [PubMed Central ID: PMC6883034]. https://doi.org/10.1038/s41419-019-2145-5.
-
24.
Zhou Z, Sun B, Nie A, Yu D, Bian M. Roles of aminoacyl-tRNA synthetases in cancer. Front Cell Dev Biol. 2020;8:599765. [PubMed ID: 33330488]. [PubMed Central ID: PMC7729087]. https://doi.org/10.3389/fcell.2020.599765.
-
25.
Ahn YH, Oh SC, Zhou S, Kim TD. Tryptophanyl-tRNA synthetase as a potential therapeutic target. Int J Mol Sci. 2021;22(9). [PubMed ID: 33926067]. [PubMed Central ID: PMC8123658]. https://doi.org/10.3390/ijms22094523.
-
26.
Francklyn CS, Mullen P. Progress and challenges in aminoacyl-tRNA synthetase-based therapeutics. J Biol Chem. 2019;294(14):5365-85. [PubMed ID: 30670594]. [PubMed Central ID: PMC6462538]. https://doi.org/10.1074/jbc.REV118.002956.
-
27.
Kim SH, Bae S, Song M. Recent development of aminoacyl-tRNA synthetase inhibitors for human diseases: A future perspective. Biomolecules. 2020;10(12). [PubMed ID: 33271945]. [PubMed Central ID: PMC7760260]. https://doi.org/10.3390/biom10121625.
-
28.
Kim T, Lee K, Nahm JH, Kim EY, Lee SH, Chang YS. Methionyl-tRNA synthetase and aminoacyl-tRNA synthetases interacting multi-functional protein-lacking exon 2 as potential diagnostic biomarkers for lung cancer. Am J Cancer Res. 2021;11(6):3135-44. [PubMed ID: 34249450]. [PubMed Central ID: PMC8263685].
-
29.
Tjong E, Dimri M, Mohiuddin SS. Biochemistry, tetrahydrofolate. StatPearls. Treasure Island (FL): StatPearls Publishing Copyright © 2023, StatPearls Publishing LLC; 2023.
-
30.
Zeleznik OA, Eliassen AH, Kraft P, Poole EM, Rosner BA, Jeanfavre S, et al. A prospective analysis of circulating plasma metabolites associated with ovarian cancer risk. Cancer Res. 2020;80(6):1357-67. [PubMed ID: 31969373]. [PubMed Central ID: PMC7073287]. https://doi.org/10.1158/0008-5472.CAN-19-2567.
-
31.
Radisauskas R, Kuzmickiene I, Milinaviciene E, Everatt R. Hypertension, serum lipids and cancer risk: A review of epidemiological evidence. Medicina (Kaunas). 2016;52(2):89-98. [PubMed ID: 27170481]. https://doi.org/10.1016/j.medici.2016.03.002.
-
32.
Ulmer H, Borena W, Rapp K, Klenk J, Strasak A, Diem G, et al. Serum triglyceride concentrations and cancer risk in a large cohort study in Austria. Br J Cancer. 2009;101(7):1202-6. [PubMed ID: 19690552]. [PubMed Central ID: PMC2768093]. https://doi.org/10.1038/sj.bjc.6605264.
-
33.
Lee DY, Lee TS. Associations between metabolic syndrome and gynecologic cancer. Obstet Gynecol Sci. 2020;63(3):215-24. [PubMed ID: 32489965]. [PubMed Central ID: PMC7231948]. https://doi.org/10.5468/ogs.2020.63.3.215.
-
34.
Garg G, Yilmaz A, Kumar P, Turkoglu O, Mutch DG, Powell MA, et al. Targeted metabolomic profiling of low and high grade serous epithelial ovarian cancer tissues: a pilot study. Metabolomics. 2018;14(12):154. [PubMed ID: 30830441]. https://doi.org/10.1007/s11306-018-1448-3.
-
35.
Kiskova T, Kassayova M. Resveratrol action on lipid metabolism in cancer. Int J Mol Sci. 2019;20(11). [PubMed ID: 31159437]. [PubMed Central ID: PMC6601040]. https://doi.org/10.3390/ijms20112704.
-
36.
Yu XH, Ren XH, Liang XH, Tang YL. Roles of fatty acid metabolism in tumourigenesis: Beyond providing nutrition (Review). Mol Med Rep. 2018;18(6):5307-16. [PubMed ID: 30365095]. https://doi.org/10.3892/mmr.2018.9577.
-
37.
Visweswaran M, Arfuso F, Warrier S, Dharmarajan A. Aberrant lipid metabolism as an emerging therapeutic strategy to target cancer stem cells. Stem Cells. 2020;38(1):6-14. [PubMed ID: 31648395]. https://doi.org/10.1002/stem.3101.
-
38.
Konstorum A, Lynch ML, Torti SV, Torti FM, Laubenbacher RC. A systems biology approach to understanding the pathophysiology of high-grade serous ovarian cancer: Focus on Iron and fatty acid metabolism. OMICS. 2018;22(7):502-13. [PubMed ID: 30004845]. [PubMed Central ID: PMC6059353]. https://doi.org/10.1089/omi.2018.0060.
-
39.
Amezaga J, Arranz S, Urruticoechea A, Ugartemendia G, Larraioz A, Louka M, et al. Altered red blood cell membrane fatty acid profile in cancer patients. Nutrients. 2018;10(12). [PubMed ID: 30513730]. [PubMed Central ID: PMC6315925]. https://doi.org/10.3390/nu10121853.
-
40.
Koundouros N, Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br J Cancer. 2020;122(1):4-22. [PubMed ID: 31819192]. [PubMed Central ID: PMC6964678]. https://doi.org/10.1038/s41416-019-0650-z.
-
41.
Eldor J. Linoleic acid: the code of life. J Health Sci Dev. 2018;1(2):18-32.
-
42.
Kiesel VA, Sheeley MP, Coleman MF, Cotul EK, Donkin SS, Hursting SD, et al. Pyruvate carboxylase and cancer progression. Cancer Metab. 2021;9(1):20. [PubMed ID: 33931119]. [PubMed Central ID: PMC8088034]. https://doi.org/10.1186/s40170-021-00256-7.
-
43.
Lao-On U, Attwood PV, Jitrapakdee S. Roles of pyruvate carboxylase in human diseases: from diabetes to cancers and infection. J Mol Med (Berl). 2018;96(3-4):237-47. [PubMed ID: 29362846]. https://doi.org/10.1007/s00109-018-1622-0.