Abstract
Background:
Nowadays, due to the increasing number of immunocompromised patients, mycosis infections caused by Candida species are on the rise, especially among hospitalized patients.Objectives:
In the current study, the chromogenic medium CHROMagar™ Candida and semi-nested polymerase chain reaction (snPCR) were compared concerning their ability to detect the species of Candida in 65 clinical isolates.Materials and Methods:
We used snPCR with universal and species-specific primers to detect Candida species in the culture of clinical isolates. By using universal primers, we carried out the amplification of the 3/end of 5.8S ribosomal DNA (rDNA) and the 5/end of 28S rDNA, including the internal transcribed spacer 2 (ITS2) and production of 350 to 410-bp fragments from 4 Candida species, vis. Candida albicans, Candida tropicalis, Candida glabrata, and Candida parapsilosis.Results:
The phenotypic identification system identified 60 (92.3%) yeasts isolates, including C. albicans (n = 33), C. glabrata (n = 14), C. tropicalis (n = 11), and C. parapsilosis (n = 2), and 5 isolates were mixed culture. By snPCR, 63 (96.6%) isolates were identified, including C. albicans (n = 37), C. glabrata (n = 11), C. tropicalis (n = 14), and C. parapsilosis (n = 1), and the species of 2 isolates could not be identified. Additionally, snPCR for the specific identification of the Candida species of the 65 clinical Candida isolates revealed 70.3% results agreement with the chromogenic medium CHROMagar™ Candida.Conclusions:
In this study, snPCR was specific and more sensitive than the chromogenic medium CHROMagar™ Candida for the detection of Candida spp. insofar as it failed to identify only a few isolates.Keywords
1. Background
Candida species are the normal flora of the mucosal surfaces of the oral cavity, gastrointestinal tract, and genitalia in humans and animals. However, they may be responsible for the infection of the mucosa of the oropharynx and vulva and vagina. Candida spp. also play a role in systemic infections in the presence of risk factors such as diabetes mellitus, AIDS, and other immunosuppressive conditions (1). The most commonly isolated species include Candida albicans, Candida glabrata, Candida tropicalis, and Candida parapsilosis. A study from Turkey (2) reported that C. albicans and C. parapsilosis were the most common agents isolated from pediatric/neonatal patients with invasive candidiasis. The rapid identification of Candida species is necessary for a timely diagnosis and selection of a suitable antifungal therapy (3-6).
The aim of the present study was to compare the molecular method semi-nested polymerase chain reaction (snPCR) with the conventional methods for the identification of the clinical isolates of Candida spp. A previous study reported that snPCR detected Candida species (C. albicans, C. tropicalis, C. glabrata, and C. parapsilosis) in a short period of time, with high sensitivity and specificity by using universal and species-specific primers (7). The species-specific primers qualified were used for the species-specific detection of PCR-amplified ribosomal DNAs (rDNAs) for 4 Candida species, vis. C. albicans, C. parapsilosis, C. tropicalis, and C. glabrata.
The method of snPCR, as reported by a previous researcher (7), utilizes universal fungal primers and a section of the 5.8S and 28S rDNAs containing the intervention of an internal transcribed spacer (ITS2). It is amplified and reamplified in snPCR using species-specific primers supplementary to the unique sequences within the ITS2 together with a common fungal primer.
2. Objectives
The aim of this investigation was to compare the use of chromogenic medium CHROMagar™ Candida and reaction of snPCR for the identification of Candida species.
3. Materials and Methods
Sixty-five clinical specimens of Candida species isolated from urine, oral mucosa, nail, and skin were assayed by snPCR. The sources of these Candida isolates are shown in Table 1.
Clinical Candida Isolates Used
| Source of Isolation | No. of Isolates |
|---|---|
| Oral mucosa | 60 |
| Urine | 3 |
| Nail | 1 |
| Skin | 1 |
| Total | 65 |
3.1. Culture
All the isolated clinical specimens of Candida spp. were subcultured on Sabouraud dextrose agar (Merck, Germany) at 37°C for 48 hours. The samples were stored in water distilled at -20°C for future assays.
3.2. Identification
All the yeast isolates were identified by wet mount, CHROMagar™ Candida medium (CHROMagar™, Paris, France), for the colony evaluation of color and phenotypic identification, including germ tube formation in bovine serum at 37°C for 3 hours, clamidoconidia formation growth in corn meal agar plus Tween 80 at 30°C for 48 hours, and growth at 45°C according to previous studies (8-10). For the molecular identification of the Candida species, snPCR was performed using universal and species-specific oligonucleotide primers.
3.3. DNA Extraction
From broth cultures, genomic Candida DNA was extracted using the glass-bead (0.2 μ) disruption (11, 12). First, a loop of fresh yeast was added to 300 mg of glass-beads (0.5 mm diameter) plus 300 μL of lysis buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, and 1% SDS) and vortexed vigorously 5 - 10 minutes for crushing the cell walls of the yeast completely. Then, it was boiled at 100°C for 20 minutes, mixed with 150 μL of 3 M sodium acetate, kept at -20°C for 10 minutes, and centrifuged at 12000 rpm for 10 minutes. The supernatant was extracted by mixing it with the same volume of phenol: chloroform: isoamyl alcohol (25:24:1), short vortexing, and centrifuging at 12000 rpm for 10 minutes. The supernatant was discarded and mixed with the same volume of chloroform, short vortexed, and centrifuged at 12000 rpm for 10 minutes. The supernatant (DNA) was precipitated by adding an equal volume of 2-propanol at -20°C for 10 minutes and then centrifuged at 12000 rpm for 10 min. Moreover, 2-propanol was discarded and the DNA was washed with 150 (300) μL of 70% ethanol at 12000 rpm for 10 minutes. (note that in this step, the tubes were not vortexed.) Ethanol was discarded, and the DNA was air-dried and resuspended in 50 μL of sterile deionized water (ddH2O) and kept at -20°C until use (12). Aliquot of 1 μL of the product solution was used as template in the PCR reaction.
3.4. Semi-Nested PCR and Primers Used
In the first amplification step, we used the universal primers of a 22-bp forward primer CTSF (5’-TCGCATCGATGAAGAACGCAGC-3’) and a 25-bp reverse primer CTSR (5’-TCTTTTCCTCCGCTTATTGATATGC-3’) (CinnaGen Inc., Iran). The ability to amplify the end of 3’end of rDNA and 5.8S the 5’ end of 28S rDNAs, including the intervening ITS2region, was tested. Species-specific oligonucleotide primers for snPCR were derived from the ITS2 regions of Candida albicans (CADET, 5’-ATTGCTTGCGGCGGTAACGTCC-3’), Candidaparapsilosis (CPDET, 5’-ACAAACTCCAAAACTTCTTCCA-3’), Candida tropicalis (CTDET, 5’-AACGCTTATTTTGCTAGTGGCC-3’), and Candida glabrata (CGDET, 5’-TAGGTTTTACCAACTCGGTGTT-3’), as described by previous studies (7, 13).
3.5. Amplification of PCR and Detection
The molecular identification of the 65 clinical isolates was carried out by snPCR according to Ahmad et al. (7). The steps of 1 and 2 of snPCR were carried out in the same conditions except for primers and cycling numbers, which were conducted according to another previous study (6). The amplification of Candida DNA was carried out in an Eppendorf Mastercycler® gradient thermocycler. Both the first and second steps of PCR were carried out in a volume of 25 μL. The PCR mixture of the first step, containing 2.5 μL of 10X PCR buffer, 0.5 μL of 10 pmol of each of CTSF and CTSR primers, 0.5 μL of 1.5 mM MgCl2, 0.5 μL of 10 mM dNTP, 0.35 μL of Tag DNA polymerase (0.625 U) (CinnaGen Inc., Iran), 3 μL of extracted DNA as template, and 17.15 μL of autoclaved ddH2O, was added. Three μL of the products of the first step of snPCR was amplified as template in the second step under the same PCR mixture and programs. Forward ITS1 primer (CTS1F) was replaced with species-specific forward primers (CADET, CPDET, CGDET, and CTDET) in 4 separate tubes and the reverse primer (CTS1R) was also used. The amplification conditions for steps 1 and 2 included one cycle denaturation at 94°C for 1 minute, annealing at 60°C for 30 sec, and extension at 72°C for 1 minute. An initial denaturation step at 94°C for 3 minutes and a final extension step at 72°C for 10 minutes were also conducted. Amplification was determined with 30 cycles in the first step and 20 cycles in the second step of snPCR. For the detection of the amplified DNA fragments, 8.5 μL of the PCR product mixed with 1.5 loading dye was loaded onto 2% agarose gel containing 0.5 μg.mL-1 ethidium bromide. Electrophoresis was thereafter carried out. The gels were exposed to UV light at 100 V for 45 minutes and then photographed. The sizes of the amplified DNA fragments were identified by comparison with molecular size marker DNA (100-bp DNA ladder).
4. Results
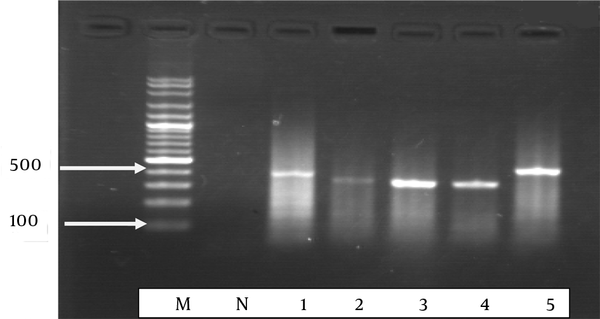
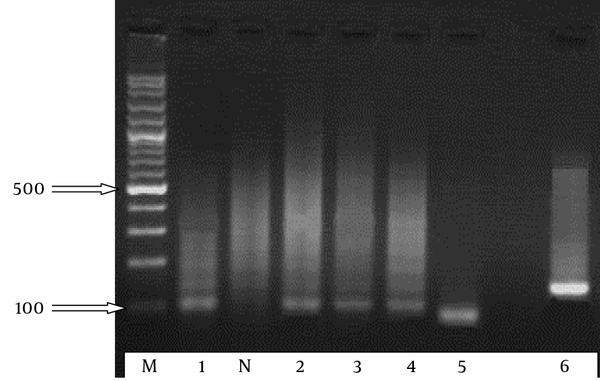
From the 65 clinical Candida isolates, CHROMagar™ Candida and phenotypic identification system identified 60 (92.3%) yeasts isolates. Candidaalbicans was the most common species (n = 33), followed by C.glabrata (n = 14), C.tropicalis (n = 11), and C.parapsilosis (n = 2), and 5 isolates were mixed culture. The snPCR amplification of the rDNAs of the 4 Candida species, viz. C.albicans, C.glabrata, C.tropicalis, and C.parapsilosis, was carried out using universal fungal primers (CTSF and CTSR). From the reamplification of the product of the first PCR with CTSR and the species-specific primers arising from the ITS2 regions of C.albicans, C.parapsilosis, C.tropicalis, and C.glabrata, different sizes of the amplified DNA fragments were obtained. For example, CTSR plus CGDET amplified a product with a size of ~ 140 bp in snPCR when the first reaction of snPCR was carried out with the template DNA from C. glabrata. The snPCR amplification of the DNAs from CTSR with primers CADET, CPDET, and CTDET amplified the products with molecular size markers of approximately 122, 100, and 130 bp, respectively. By snPCR, 63 (96.6%) isolates were identified, including C.albicans (n = 37), C.glabrata (n = 11), C.tropicalis (n = 14), and C. parapsilosis (n = 1). Also, snPCR for the specific identification of the Candida species of the 65 clinical Candida isolates revealed that the percent agreement was 70.3% between the positive results of 63 (92.9%) of snPCR and 60 (92.3%) of CHROMagar™ Candida and phenotypic results (Table 2). In addition, the results of 6 samples obtained using CHROMagar™ did not according to snPCR and the species of 2 isolates could not be identified by snPCR (Table 3). For the identification of the quantities of DNA, the optical densities of the DNAs were measured in 260 nm for DNA and 280 nm for proteins. The ratio of 260/280 nm of the tested samples was obtained between 1.5 and 2.0.
Identification of Clinical Candida Isolates by CHROMagar™ and Phenotypic Methods and Agreement With SnPCR
| Clinical Isolates | CHROMagar™ and Phenotypic | SnPCR |
|---|---|---|
| U13N | Candida parapsilosis | Candida glabrata |
| U11N | Candida glabrata | Candida tropicalis |
| OC 36, OC 37,44 | Candida albicans | Candida glabrata |
| OC 45, 46,48, 54 | Candida glabrata | Candida albicans |
| OC 32/1, 73/1 | Mixed (Candida albicans and Candida tropicalis) | Candida tropicalis |
| OC 7, 95, 69/1 | Mixed (Candida albicans and Candida tropicalis) | Candida tropicalis |
| OC 6, 30 | Candida albicans | NI |
Semi-Nested PCR 1
Semi-Nested PCR 2
5. Discussion
Nowadays, despite progresses in therapeutic methods and sanitary cares, the incidence of invasive systemic mycoses, especially opportunistic fungal pathogens such as Candida, is on the increase (14). Although C.albicans is the most frequent cause of candidiasis, the incidence of the disease caused by other species of Candida has been increasing drastically (11, 14). Among the azoles, fluconazole has revealed tolerance; and antifungal drug resistance is becoming a major problem, especially in immunocompromised patients (15, 16). Resistance has been shown to Candida krusei and Candida glabrata (17). Therefore, a rapid and accurate diagnosis of the species may assist in finding an appropriate therapy in the cases of candidiasis and thus reduce the frequency of isolates resistant to antifungal agents such as azoles, which have therapeutic activity against Candida species. In this study, the chromogenic medium CHROMagar™ Candida and snPCR were compared regarding their ability to recognize the species of 65 clinical isolates of Candida spp. The latter method is a strong method for amplifying small amounts of DNA, but it becomes disputable when it is used for the detection of more than 1 species (18). Moreover, snPCR has been demonstrated to be a useful, rapid diagnostic tool for the detection of panfungal genome (19). In the current investigation, snPCR experiments targeting species-specific sequences in the rDNA were begun for the specific recognition of 4 Candida species by using reference strains. The aim for snPCR amplification was rDNA. Although PCR experiments with other target sequences have been reported in the previous studies (20, 21), the use of rDNA for a sensitive recognition of Candida has been observed to be the most competent because it is present in numerous copies (50 to 100 copies) per Candida genome (20). Also, PCR experiments with numerous copy purposes are more sensitive than those with single-copy purposes (19, 20). Furthermore, in the middle of the preserved rDNA subunits are the internal transcribed spacers, which include sequences matchless to each Candida sp., and therefore the use of primers according to these regions facilitates species recognition (6). Multiplex PCR assays have detected Candida scpecies according to previous studies (22, 23). Nevertheless, for a positive result, multiplex PCRs require a minimum of 20 cells, compared to 0.2 cells for snPCR. Therefore, the snPCR assay is at least 100 times more sensitive than multiplex PCRs. According to our findings, PCR with species-specific primers that amplify genes ITS1 and ITS2 for rDNA is a very specific and sensitive method for the recognition of C. glabrata and C. albicans, and with less specificity for C. tropicalis; these results are concordant with some previous observations (24). Previous studies have reported that commercial techniques for the recognition of Candida species misrecognized some clinical Candida isolates (25-29). Also, several other studies have revealed that PCR is more sensitive than conventional culture methods in the diagnosis of candidiasis (30-32).
In conclusion, our results demonstrated that snPCR is a specific and sensitive method for the diagnosis of candidemia caused by 4 clinically important Candida species, viz. Candida albicans, Candida parapsilosis, Candida tropicalis, and Candida glabrata. Furthermore, snPCR is a rapid method for the recognition of patients infected with more than one Candida species. Needless to say; a rapid diagnosis will contribute to the choice of suitable therapeutic agents.
Acknowledgements
References
-
1.
Weig M, Gross U, Muhlschlegel F. Clinical aspects and pathogenesis of Candida infection. Trends Microbiol. 1998;6(12):468-70. [PubMed ID: 10036723].
-
2.
Kuzucu C, Durmaz R, Otlu B, Aktas E, Gulcan H, Cizmeci Z. Species distribution, antifungal susceptibility and clonal relatedness of Candida isolates from patients in neonatal and pediatric intensive care units at a medical center in Turkey. New Microbiol. 2008;31(3):401-8. [PubMed ID: 18843896].
-
3.
Cuenca-Estrella M, Rodriguez D, Almirante B, Morgan J, Planes AM, Almela M, et al. In vitro susceptibilities of bloodstream isolates of Candida species to six antifungal agents: results from a population-based active surveillance programme, Barcelona, Spain, 2002-2003. J Antimicrob Chemother. 2005;55(2):194-9. [PubMed ID: 15618284]. https://doi.org/10.1093/jac/dkh548.
-
4.
Foster N, Symes C, Barton R, Hobson R. Rapid identification of Candida glabrata in Candida bloodstream infections. J Med Microbiol. 2007;56(Pt 12):1639-43. [PubMed ID: 18033833]. https://doi.org/10.1099/jmm.0.47406-0.
-
5.
Nguyen MH, Peacock JJ, Morris AJ, Tanner DC, Nguyen ML, Snydman DR, et al. The changing face of candidemia: emergence of non-Candida albicans species and antifungal resistance. Am J Med. 1996;100(6):617-23. [PubMed ID: 8678081].
-
6.
Pfaller MA, Jones RN, Doern GV, Sader HS, Messer SA, Houston A, et al. Bloodstream Infections Due to Candida Species: SENTRY Antimicrobial Surveillance Program in North America and Latin America, 1997-1998. Antimicrobial Agents and Chemotherapy. 2000;44(3):747-51. https://doi.org/10.1128/aac.44.3.747-751.2000.
-
7.
Ahmad S, Khan Z, Mustafa AS, Khan ZU. Seminested PCR for diagnosis of candidemia: comparison with culture, antigen detection, and biochemical methods for species identification. J Clin Microbiol. 2002;40(7):2483-9. [PubMed ID: 12089267].
-
8.
Sadeghi Nejad B, Rafiei A, Moosanejad F. Prevalence of Candida species in the oral cavity of patients with periodentitis. African J of Biotechnol. 2013;10(15):2987-90.
-
9.
Sandven P. Laboratory identification and sensitivity testing of yeast isolates. Acta Odontol Scand. 1990;48(1):27-36. [PubMed ID: 1690944].
-
10.
Williams DW, Lewis MA. Isolation and identification of Candida from the oral cavity. Oral Dis. 2000;6(1):3-11. [PubMed ID: 10673781].
-
11.
Mirhendi H, Makimura K, Zomorodian K, Maeda N, Ohshima T, Yamaguchi H. Differentiation of Candida albicans and Candida dubliniensis using a single-enzyme PCR-RFLP method. Jpn J Infect Dis. 2005;58(4):235-7. [PubMed ID: 16116258].
-
12.
Yamada Y, Makimura K, Merhendi H, Ueda K, Nishiyama Y, Yamaguchi H, et al. Comparison of different methods for extraction of mitochondrial DNA from human pathogenic yeasts. Jpn J Infect Dis. 2002;55(4):122-5. [PubMed ID: 12403909].
-
13.
Fujita S, Lasker BA, Lott TJ, Reiss E, Morrison CJ. Microtitration plate enzyme immunoassay to detect PCR-amplified DNA from Candida species in blood. J Clin Microbiol. 1995;33(4):962-7. [PubMed ID: 7790469].
-
14.
Neppelenbroek KH, Campanha NH, Spolidorio DM, Spolidorio LC, Seo RS, Pavarina AC. Molecular fingerprinting methods for the discrimination between C. albicans and C. dubliniensis. Oral Dis. 2006;12(3):242-53. [PubMed ID: 16700733]. https://doi.org/10.1111/j.1601-0825.2005.01189.x.
-
15.
Barada G, Basma R, Khalaf RA. Microsatellite DNA identification and genotyping of Candida albicans from Lebanese clinical isolates. Mycopathologia. 2008;165(3):115-25. [PubMed ID: 18266073]. https://doi.org/10.1007/s11046-008-9089-0.
-
16.
Xu J, Ramos AR, Vilgalys R, Mitchell TG. Clonal and spontaneous origins of fluconazole resistance in Candida albicans. J Clin Microbiol. 2000;38(3):1214-20. [PubMed ID: 10699025].
-
17.
Cirak MY, Kalkanci A, Kustimur S. Use of molecular methods in identification of Candida species and evaluation of fluconazole resistance. Mem Inst Oswaldo Cruz. 2003;98(8):1027-32. [PubMed ID: 15049085].
-
18.
Than LT, Chong PP, Ng KP, Seow HF. Detection of 10 medically important Candida species by seminested polymerase chain reaction. Diagn Microbiol Infect Dis. 2012;72(2):196-8. [PubMed ID: 22154674]. https://doi.org/10.1016/j.diagmicrobio.2011.10.008.
-
19.
Bagyalakshmi R, Therese KL, Madhavan HN. Application of semi-nested polymerase chain reaction targeting internal transcribed spacer region for rapid detection of panfungal genome directly from ocular specimens. Indian j of ophthalm. 2007;55(4):261.
-
20.
Mitchell TG, Sandin RL, Bowman BH, Meyer W, Merz WG. Molecular mycology: DNA probes and applications of PCR technology. J Med Vet Mycol. 1994;32 Suppl 1:351-66. [PubMed ID: 7722798].
-
21.
Reiss E, Tanaka K, Bruker G, Chazalet V, Coleman D, Debeaupuis JP, et al. Molecular diagnosis and epidemiology of fungal infections. Med Mycol. 1998;36 Suppl 1:249-57. [PubMed ID: 9988514].
-
22.
Chang HC, Leaw SN, Huang AH, Wu TL, Chang TC. Rapid identification of yeasts in positive blood cultures by a multiplex PCR method. J Clin Microbiol. 2001;39(10):3466-71. [PubMed ID: 11574557]. https://doi.org/10.1128/JCM.39.10.3466-3471.2001.
-
23.
Fujita SI, Senda Y, Nakaguchi S, Hashimoto T. Multiplex PCR using internal transcribed spacer 1 and 2 regions for rapid detection and identification of yeast strains. J Clin Microbiol. 2001;39(10):3617-22. [PubMed ID: 11574582]. https://doi.org/10.1128/JCM.39.10.3617-3622.2001.
-
24.
Oliveira NDC, Pontello Rampazzo RDC, Minari MC, Correa PRC, Bizerra FC, Carneiro M, et al. Use of chromogenic medium and semi-nested PCR-based assay to identify Candida species [in Portuguese]. Semina. 2006;27(2):125-32.
-
25.
Crist AJ, Johnson LM, Burke PJ. Evaluation of the Microbial Identification System for identification of clinically isolated yeasts. J Clin Microbiol. 1996;34(10):2408-10. [PubMed ID: 8880489].
-
26.
Espinel-Ingroff A, Stockman L, Roberts G, Pincus D, Pollack J, Marler J. Comparison of RapID yeast plus system with API 20C system for identification of common, new, and emerging yeast pathogens. J Clin Microbiol. 1998;36(4):883-6. [PubMed ID: 9542903].
-
27.
Fenn JP, Segal H, Barland B, Denton D, Whisenant J, Chun H, et al. Comparison of updated Vitek Yeast Biochemical Card and API 20C yeast identification systems. J Clin Microbiol. 1994;32(5):1184-7. [PubMed ID: 8051243].
-
28.
Lo HJ, Ho YA, Ho M. Factors accounting for misidentification of Candida species. J Microbiol Immunol Infect. 2001;34(3):171-7. [PubMed ID: 11605807].
-
29.
Verweij PE, Breuker IM, Rijs AJ, Meis JF. Comparative study of seven commercial yeast identification systems. J Clin Pathol. 1999;52(4):271-3. [PubMed ID: 10474518].
-
30.
Chryssanthou E, Andersson B, Petrini B, Lofdahl S, Tollemar J. Detection of Candida albicans DNA in serum by polymerase chain reaction. Scand J Infect Dis. 1994;26(4):479-85. [PubMed ID: 7984982].
-
31.
Khan ZU, Mustafa AS. Detection of Candida species by polymerase chain reaction (PCR) in blood samples of experimentally infected mice and patients with suspected candidemia. Microbiol Res. 2001;156(1):95-102. [PubMed ID: 11372661]. https://doi.org/10.1078/0944-5013-00072.
-
32.
Wahyuningsih R, Freisleben HJ, Sonntag HG, Schnitzler P. Simple and rapid detection of Candida albicans DNA in serum by PCR for diagnosis of invasive candidiasis. J Clin Microbiol. 2000;38(8):3016-21. [PubMed ID: 10921970].