Abstract
Background:
Apoptosis is a crucial process in the failure of cancer progression. However, the occurrence of resistance to chemotherapy in cancer cells may prevent apoptosis via induction of the expression of the anti-apoptotic genes (Bcl-2) and/or reduction of the expression of the apoptotic caspases.Objectives:
The current study aimed at investigating the apoptotic effects of targeted co-delivery of docetaxel and cMet siRNA (siMet) through mucin1 aptamer-conjugated chitosan nanoparticles on mucin1 + metastatic breast cancer cells (SKBR3).Methods:
Characterization of nano-drugs, Western blotting assay, annexin V/ propidium iodide staining assay, and gene expression studies were evaluated based on metastatic breast cancer cells.Results:
Characterization showed the appropriate size (110.5 ± 3.9 nm), zeta potential (11.6 ± 0.8 mV), and spherical shape of nanoparticles. Loading efficiency of 90.7% and 88.3% were gained for siRNA and docetaxel, respectively. The siRNA entrapment onto nanoparticles and conjugation of aptamers to nanoparticles were confirmed by gel electrophoresis. Gene knockdown assay represented the effectiveness of siMet on cMet gene silencing. According to the flow cytometry results, targeted co-delivery was successful in leading tumor cells to apoptosis (94.9%). Also, targeted co-delivery could reduce the expression of Bcl-2 gene (P < 0.0001) and increase the expression of caspase-8 and caspase-9 genes (P < 0.0001).Conclusions:
Combination treatment of metastatic breast cancer cells with aptamer-conjugated nanoparticles containing docetaxel and cMet siRNA significantly increased apoptosis.Keywords
Apoptosis Docetaxel Metastatic Breast Cancer Mucin1 Aptamer siRNA
1. Background
Apoptosis, the programmed cell death, is one of the vital pillars of combating metastatic breast cancer. Reduced apoptosis or escaping of the tumor cells from apoptosis causes cancer progression (1) and may lead to metastasis (2). Apoptosis is classified into two extrinsic and intrinsic pathways. The interaction of distinct transmembrane receptors with their ligands activates extrinsic pathway, which leads to activation of caspase-8 (Cas-8). The Bcl-2 protein family is responsible for the initiation of the intrinsic pathway that causes the formation of apoptosome complex and ultimately the activation of caspase-9 (Cas-9) enzyme (3). The c-Met, the tyrosine kinase receptor, and its ligand hepatocyte growth factor (HGF) are the crucial proteins for the growth of epithelial cells (4). The overexpression of c-Met is related to resistance to apoptosis in various cancer cells. Also, downregulation of c-Met potentiates different aspects of apoptotic pathways such as Fas-mediated apoptosis or Bcl-2 downregulation (5-7).
Docetaxel (DTX), one of the taxanes in standard treatment of breast cancer, acts through inhibition of microtubular depolymerization. Also, DTX induces apoptosis via the mechanisms such as increasing TNF-α production in tumor cells (8) and resistance to DTX leads to decreased apoptosis of breast cancer cells (9). In addition to chemotherapy, application of gene silencing tools such as short-interfering RNA (siRNA) is an interesting field of research. One of the procedures to increase delivery of negatively charged, fast degradable siRNA to the cancer cells is the utilization of positively charged, biocompatible, and non-immunogenic chitosan nanoparticles (NPs) (10). In order to design a targeted delivery system, aptamer (APT) molecules are introduced. APTs are the synthetic, safe, non-immunogenic, and specific oligonucleotides bind to their target with high affinity (11). Mucin1 is one of the tumor-associated antigens that is aberrantly overexpressed in breast cancer cells and, therefore, can be a desired target for APT (12).
2. Objectives
The current study designed a novel delivery system of mucin1 APT-conjugated chitosan NPs containing DTX and c-Met siRNA (siMet). The apoptotic effects of this system were evaluated on mucin1 + metastatic breast cancer cells (SKBR3) by annexin V-FITC /propidium iodide (PI) apoptosis assay. Also, the expression of the important genes (Bcl-2, Cas-8, and Cas-9) involved in extrinsic or intrinsic pathways of apoptosis was assessed.
3. Methods
3.1. Materials
The SKBR3 was supplied by the National Cell Bank of Iran (Pasteur Institute of Iran). The Roswell Park Memorial Institute (RPMI) 1640 medium, fetal bovine serum (FBS), trypsin, and penicillin-streptomycin were obtained from Gibco (USA). The 50-kDa depolymerized chitosan (CS) was a gift from Prof. Fatemeh Atyabi (Nanotechnology Research Centre, Tehran University of Medical Sciences). Carboxymethyl dextran (CMD), N-hydroxysuccinimide (NHS), 1-ethyl-3-(3-dimethylaminopropyl), and carbodiimide (EDC) were purchased from Merck (Germany). Human-specific cMET-siRNA (Catalog number: sc-29397) and the non-specific siRNA (Catalog number: sc-37007) were supplied by Santa Cruz Biotechnology (USA). DTX was purchsed from AqVida (Germany). The APT with the sequence of 5’ -amino-C6-GGG AGA CAA GAA TAA ACG CTC AAG AAG TGA AAA TGA CAG AAC ACA ACA TTC GAC AGG AGG CTC ACA ACA GGC- 3’ was synthesized by TAG Copenhagen (Denmark). RNA extraction solution (RNX-Plus®) was purchased from SinaClon (Iran). RIPA lysis buffer system, cMET primary antibody, and horseradish-conjugated anti-IgG secondary antibody were supplied by Santa Cruz Biotechnology (USA). The protein assay kit was supplied by Razibiotech (Iran). Electrochemiluminescent substrate was obtained from Thermo Fisher Scientific (USA). FITC Annexin V Apoptosis Detection Kit I was purchased from BD Biosciences (USA).
3.2. Pharmaceutical Characterizations
3.2.1. Preparation of Pharmaceutical Compound
Loaded nanoparticles (NPs) were prepared by dissolving CS (0.1% w/v) in diethylpyrocarbonate (DEPC)-treated water with a magnetic stirrer for two hours. Then the siMet (57 μg/3μL) and DTX (0.25 µg/5µL) were slowly added to 1.2 mL of CMD (0.1% w/v) solution. Thereafter, the prepared mixture was dropwisely added into 1 mL of CS solution and incubated for 30 minutes at 25ºC. Mucin1 APT was conjugated to NPs according to the previously described method (13).
3.2.2. Physicochemical Characteristics of Nanoparticles
The particle size, zeta potential, and polydispersity index (PDI) were assessed with Photon Correlation Spectroscopy using Zetasizer Nano-ZS (Malvern Instruments Ltd, Malvern, Worcestershire, UK) at the wavelength of 633 nm. Transmission electron microscope (TEM, LE-O906 Zeiss®, Oberkochen, Germany) was used to evaluate the morphological properties of freshly prepared nanoparticles.
3.2.3. Loading Efficiency
The loading efficiency (LE) of DTX and siMet was evaluated with UV-vis spectrophotometer (Nanodrop® 2000, Thermo Scientific, Worcester, MA, USA). DTX-loaded NPs were centrifuged at 14,000 rpm for 30 minutes and the optical density (OD) of the free concentrated DTX in the supernatant was read in triplicate at 230 nm. Also, the OD of free siMet was read in triplicate at 260 nm after centrifugation of siMet-loaded NPs at 11,000 rpm for 20 minutes. The OD of the supernatant regained from unloaded NPs was read as the blank. The following equation represents the LE% calculation:
LE (%) = [1 - (OD of free DTX or siRNA in supernatant/OD of initial feeding amount of sample)] × 100
3.2.4. Electrophoresis for siRNA Entrapment onto NPs and Confirmation of APT-NPs Conjugation
The electrophoresis in Tris-acetate-ethylenediaminetetraacetic acid (TAE buffer, 1M) solution on a 2% agarose gel was run with siMet-loaded NPs, control NPs, and naked siMet to confirm siRNA entrapment onto NPs. Also, another electrophoresis was run to confirm APT conjugation to NPs with three pharmaceutical groups including intact APT, the unpurified, and purified conjugates of NPs + APT.
3.3. Protein Extraction and Western Blotting
The SKBR3 cells were seeded (5 × 106 cell/flask) and treated with NPs + siMet for 48 hours. The RIPA lysis buffer system (Santa Cruz, USA) was used to extract the protein. The concentration of extracted proteins was determined by the Bradford method (Protein Assay Kit, Razibiotech, Iran) and the extractions were all stored at -80ºC. Then the electrophoresis on 8% SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) gel and the wet transfer system (Bio Rad, USA) were run for the transference of proteins onto PVDF (polyvinylidene fluoride or polyvinylidene difluoride) membrane and the membrane was blocked by TBST (Tris-buffered saline with Tween 20) buffer containing 3% BSA (bovine serum albumin) and incubated overnight at 4ºC. Thereafter, the 1:500 dilution of c-Met primary antibody (Santa Cruz, USA) was added and incubated at room temperature for one hour. The membrane was washed with TBST buffer in triplicate. The 1:2000 dilution of horseradish-conjugated anti-IgG secondary antibody (Santa Cruz, USA) was added at room temperature for two hours. The monoclonal beta-actin primary antibody (Santa Cruz, USA) at a dilution of 1:500 was used as the control. The created bands were detected by SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher scientific, USA).
3.4. Annexin V/PI Staining Assay
In order to detect apoptosis, the SKBR3 cells were seeded (5 × 105 cell/well) and treated with different pharmaceutical groups. Then the medium of each well was dried and stored. The cells were rinsed with PBS and the washing contents were stored. The pre-warmed 0.05% trypsin: EDTA solution was added to cover the cells; then the content was incubated for five minutes at 37ºC and observed under a microscope for releasing. Then 10% FBS containing medium was added to the detached cells. Finally, the apoptosis assay was performed according to the manufacturer's instructions (FITC Annexin V Apoptosis Detection Kit I, BD Pharmingen, USA).
3.5. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR
SKBR3 cells (5 × 105 cell/well) were cultured and treated for 24 and 48 hours. Subsequently, the RNA content was extracted using RNX-Plus® solution according to the manufacturer’s instructions. Then the UV-vis spectrophotometer (Nanodrop®) was used to detect the concentration of RNA samples at 260 nm. The high-quality samples were stored at -70ºC for future studies. Continuously, RNA samples were converted to complementary DNA (cDNA) as follows: 5 ng/µL of total RNA was mixed with 1 µL of random hexamer, 1 µL of oligo dT, 2 µL of dNTP mixture, 4 µL of 5X RT (reverse transcript) buffer and 0.5 µL of reverse transcriptase. The PCR primers of the desired and the housekeeping gene (GAPDH) were designed by OLIGO 7 primer analysis software (Table 1). Afterwards, the qRT-PCR was performed using SYBR Green Real-time PCR master mix (Ampliqon®, Denmark) in the total volume of 10 μL on the Roche LightCycler® 96 System set on denaturation at 94ºC for 10 minutes, 45 cycles of 10 seconds at 94ºC for amplification, 30 seconds for annealing and 10 seconds for extension at 72ºC. Finally, the melting curves were analyzed and the relative mRNA level was obtained by 2-ΔΔCt method.
Sequences of Primers
| Primer | Forward Sequence | Reverse Sequence |
|---|---|---|
| GAPDH | CAAGATCATCAGCAATGCCTCC | GCCATCACGCCACAGTTTCC |
| Bcl-2 | CATCAGGAAGGCTAGAGTTACC | CAGACATTCGGAGACCACAC |
| Cas-8 | ACCTTGTGTCTGAGCTGGTCT | GCCCACTGGTATTCCTCAGGC |
| Cas-9 | GCAGGCTCTGGATCTCGGC | GCTGCTTGCCTGTTAGTTCGC |
3.6. Statistics
GraphPad Prism 6.0 was used for statistical analysis. The results were analyzed by ANOVA and Tukey post hoc test as necessity. The results were expressed as mean ± standard deviation (SD) in the body and tables. The level of significance was considered less than 0.05.
4. Results
4.1. Pharmaceutical Characterizations
Table 2 represents the physicochemical properties of the desired pharmaceutical groups. As shown in Figure 1, the TEM imaging confirmed the smooth spherical NPs containing DTX and siRNA. The calculated loading efficiency was 90.7% for NPs + siMet and 88.3% for NPs + DTX. The electrophoresis of siRNA-loaded NPs confirmed the complete entrapment of siRNA onto the NPs (Figure 2). Confirmation of the high-yield APT-conjugated NPs is shown in Figure 3 by the distinct band of intact APT and the indistinguishable bands for unpurified and purified conjugations of NPs + APT. Also, the lack of band formation for the mentioned groups confirmed the perfect conjugation of APT to NPs.
Mean Diameter, Zeta Potential, and PDI
| Nano-Drug | Characteristic | ||
|---|---|---|---|
| Mean Diameter, nm | Zeta Potential, mV | PDI | |
| dCS + DTX + siRNA | 102.3 ± 6.7 | 13.2 ± 0.3 | 0.227 |
| dCS + APT + DTX + siRNA | 110.5 ± 3.9 | 11.6 ± 0.8 | 0.292 |
Morphologic characteristics of NPs + DTX + siMet

Electrophoresis of siRNA-loaded NPs; A) naked siRNA, B) NPs + siRNA (28.5 µg), C) NPs + siRNA (57 µg), D) NPs + siRNA (85.5 µg), and E) intact NPs

Confirmation of aptamer conjugation; A) intact Mucin1 APT, B) unpurified NPs + APT, and C) purified NPs + APT

4.2. Protein Expression Assay
The Western blot assay was performed to confirm the c-Met gene silencing by siMet. As represented in Figure 4, overexpression of c-Met protein was obvious in intact SKBR3 cells. Also, the expression of c-Met protein was silenced in the cells treated with NPs + siMet for 48 hours.
Protein expression assay; A) expression of cMET protein in SKBR3 cells, B) knock-downing of cMET protein via siRNA in SKBR3 cells, and C) expression of β-actin protein in SKBR3 cells

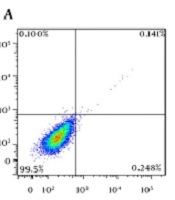
4.3. Flow Cytometry
In order to evaluate apoptotic effects of different pharmaceutical groups on SKBR3 cells, annexin V-FITC/PI staining assay was performed. According to Figure 5, the cells were divided into four types based on the results of staining as follows: (1) viable cells [annexin V (-) and PI (-)]; (2) early apoptotic cells [annexin V (+) and PI (-)]; (3) late apoptotic cells [annexin V (+) and PI (+)], and (4) necrotic cells [annexin V (-) and PI (+)]. All pharmaceutical groups significantly decreased (P < 0.0001) viable cells (%) as compared with the control group (99.5%). The majority of the cells in all treated groups were in apoptosis phase (early and late) as follows: NPs + siMet (57.9%), NPs + DTX (71.1%), NPs + siMet + DTX (80.2%), NPs + APT + siMet (87%), NPs + APT + DTX (90.4%), and NPs + APT + siMet + DTX (94.9%). Also, all pharmaceutical groups containing APT vs. corresponding APT-lacking ones significantly caused late apoptosis in SKBR3 cells (P < 0.0001).
Annexin V-FITC/PI staining assay; A) control cells, B) NPs + siMet, C) NPs + DTX, D) NPs + siMet + DTX, E) NPs + APT + siMet, F) NPs + APT + DTX, and G) NPs + APT + siMet + DTX

4.4. Gene Expression Assay
In order to evaluate the effect of different pharmaceutical groups on the expression of the genes involved in apoptosis (Bcl-2, Cas-8, and Cas-9), the qRT-PCR assay was performed and the results are represented in Figure 6. Also, the detailed results of the comparison of the groups are represented in Table 3.
Gene expression assay; A) Bcl-2 (24 h), B) Bcl-2 (48 h), C) Cas-8 (24 h), D) Cas-8 (48 h), E) Cas-9 (24 h), and F) Cas-9 (48 h)

Gene Expression Profile
| Time | Group | |||||
|---|---|---|---|---|---|---|
| NPs + siMet vs. CTRL | NPs + DTX vs. CTRL | NPs + siMet + DTX vs. CTRL | NPs + siMet vs. NPs + APT + siMet | NPs + DTX vs. NPs + APT + DTX | NPs + siMet + DTX vs. NPs + APT + siMet + DTX | |
| Bcl-2 | ||||||
| 24 h | 0.79 ± 0.03 vs. 1.0 (P < 0.0001) | 0.85 ± 0.03 vs. 1.0 (P < 0.05) | 0.59 ± 0.04 vs. 1.0 (P < 0.0001) | NS | 0.85 ± 0.03 vs. 0.31 ± 0.06 (P < 0.0001) | 0.59 ± 0.04 vs. 0.16 ± 0.05 (P < 0.0001) |
| 48 h | 0.67 ± 0.07 vs. 1.0 (P < 0.0001) | 0.73 ± 0.02 vs. 1.0 (P < 0.0001) | 0.48 ± 0.05 vs. 1.0 (P < 0.0001) | NS | 0.73 ± 0.02 vs. 0.08 ± 0.04 (P < 0.0001) | 0.48 ± 0.05 vs. 0.0008 ± 0.0003 (P < 0.0001) |
| Cas-8 | ||||||
| 24 h | 1.20 ± 0.12 vs. 1.0 (P < 0.001) | 1.40 ± 0.02 vs. 1.0 (P < 0.0001) | 1.43 ± 0.03 vs. 1.0 (P < 0.0001) | NS | NS | 1.43 ± 0.03 vs. 1.87 ± 0.03 (P < 0.0001) |
| 48 h | 1.36 ± 0.01 vs. 1.0 (P < 0.0001) | 1.64 ± 0.05 vs. 1.0 (P < 0.0001) | 1.67 ± 0.05 vs. 1.0 (P < 0.0001) | 1.36 ± 0.01 vs. 1.51 ± 0.02 (P < 0.05) | NS | 1.67 ± 0.05 vs. 2.06 ± 0.07 (P < 0.0001) |
| Cas-9 | ||||||
| 24 h | 1.41 ± 0.07 vs. 1.0 (P < 0.001) | 1.67 ± 0.03 vs. 1.0 (P < 0.0001) | 1.98 ± 0.13 vs. 1.0 (P < 0.0001) | NS | 1.67 ± 0.03 vs. 2.0.2 ± 0.10 (P < 0.01) | 1.98 ± 0.13 vs. 2.55 ± 0.15 (P < 0.0001) |
| 48 h | 1.99 ± 0.07 vs. 1.0 (P < 0.0001) | 2.45 ± 0.05 vs. 1.0 (P < 0.0001) | 3.04 ± 0.10 vs. 1.0 (P < 0.0001) | NS | 2.45 ± 0.05 vs. 3.34 ± 0.08 (P < 0.0001) | 3.04 ± 0.10 vs. 4.37 ± 0.22 (P < 0.0001) |
5. Discussion
Apoptosis is a crucial mechanism to terminate survival of cancer cells. Activation of anti-apoptotic factors such as Bcl2 protein and attenuation of apoptotic enzymes such as caspases are two mechanisms that cancer cells use to escape apoptosis (14). One of the pathways, which protect the tumor cells from apoptosis, is c-Met/HGF axis. In addition, it activates other signaling pathways leading to cancer cell growth, proliferation, and invasion (15, 16). The current study evaluated the apoptotic effects of anti-mucin1 APT-conjugated chitosan NPs containing DTX and siMet on metastatic breast cancer cells. According to the results of characterization of nanodrugs, the smooth spherical NPs with positive zeta potential and a size of less than 150 nm were formed. This size of NPs is ideal due to fewer uptakes by macrophages (17). The spherical-shaped NPs release their contents in a controlled manner into the tumor cells. The advantage of positive zeta potential of the NPs is less exposure to plasma environment and more diffusion into the tumor tissue, which causes siRNA delivery with the least exposure to plasma endonucleases (18).
According to the results of apoptosis assay and gene expression studies, the NPs containing siMet could significantly increase apoptosis in SKBR3 cells through gene expression reduction of Bcl-2 and induction of both studied caspases (Cas-8 and Cas-9). Several studies show that blockade of c-Met through monoclonal antibodies, small molecules, or gene silencing tools activates apoptosis via Bcl-2 reduction or caspases activation (19-21). On the other hand, the NPs + DTX could lead the SKBR3 cells to apoptosis through decreasing Bcl-2 and increasing Cas-8 and Cas-9 gene expression, which is in line with the findings of other studies that taxane-induced apoptosis in cancer cells is related to inactivation of Bcl-2 (22) and increased protein expression of Cas-8 and Cas-9 (23). Also, according to these studies and the current study, the increase of Cas-9 expression was more obvious than that of Cas-8.
Although the application of chemotherapeutic agents such as docetaxel is one of the successful methods of cancer treatment, the resistance occurred after a while causes the failure in therapy. In order to prevent this resistance, co-delivery of the chemotherapeutic agent and a related siRNA through appropriate nanocarriers can be an applicable solution (24-26). It is shown that co-administration of docetaxel and anti-HGF antibody increases apoptotic effects of docetaxel through induction of Cas-3 and -7 (27). The current study results confirmed these findings, since the treated cells with NPs + siMet + DTX represented more apoptosis through reduced Bcl-2 and induced Cas-8 and Cas-9 gene expression. Also, the apoptosis was at maximum level when the cells were treated with targeted co-delivery NPs + APT + siMet + DTX. Increase in the targeted delivery of both siRNA and chemotherapeutic agents is the definitive role of APT (28). Also, several studies report the positive effect of targeted co-delivery of siRNA and anticancer agent on apoptosis level of various cancer cells (29-31).
As a conclusion, in order to increase apoptotic properties of DTX on metastatic breast cancer cells, simultaneous treatment with siMet could be an effective method. On the other hand, mucin1 aptamer can be used to increase targeted delivery of the NPs containing siMet and DTX to mucin1 + SKBR3 cells. However, the efficiency of this novel cancer treatment method in apoptosis should be investigated further in other cancer cells and animal models.
References
-
1.
Wong RS. Apoptosis in cancer: From pathogenesis to treatment. J Exp Clin Cancer Res. 2011;30:87. [PubMed ID: 21943236]. [PubMed Central ID: PMC3197541]. https://doi.org/10.1186/1756-9966-30-87.
-
2.
Su Z, Yang Z, Xu Y, Chen Y, Yu Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol Cancer. 2015;14:48. [PubMed ID: 25743109]. [PubMed Central ID: PMC4343053]. https://doi.org/10.1186/s12943-015-0321-5.
-
3.
Chaabane W, User SD, El-Gazzah M, Jaksik R, Sajjadi E, Rzeszowska-Wolny J, et al. Autophagy, apoptosis, mitoptosis and necrosis: Interdependence between those pathways and effects on cancer. Arch Immunol Ther Exp (Warsz). 2013;61(1):43-58. [PubMed ID: 23229678]. https://doi.org/10.1007/s00005-012-0205-y.
-
4.
Organ SL, Tsao MS. An overview of the c-MET signaling pathway. Ther Adv Med Oncol. 2011;3(1 Suppl):S7-S19. [PubMed ID: 22128289]. [PubMed Central ID: PMC3225017]. https://doi.org/10.1177/1758834011422556.
-
5.
Gomez-Quiroz LE, Factor VM, Kaposi-Novak P, Coulouarn C, Conner EA, Thorgeirsson SS. Hepatocyte-specific c-Met deletion disrupts redox homeostasis and sensitizes to Fas-mediated apoptosis. J Biol Chem. 2008;283(21):14581-9. [PubMed ID: 18348981]. [PubMed Central ID: PMC2386934]. https://doi.org/10.1074/jbc.M707733200.
-
6.
Sun C, Liu Z, Li S, Yang C, Xue R, Xi Y, et al. Down-regulation of c-Met and Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell proliferation, migration and colony formation. Oncotarget. 2015;6(28):25533-74. [PubMed ID: 26325180]. [PubMed Central ID: PMC4694850]. https://doi.org/10.18632/oncotarget.4575.
-
7.
Tang MK, Zhou HY, Yam JW, Wong AS. c-Met overexpression contributes to the acquired apoptotic resistance of nonadherent ovarian cancer cells through a cross talk mediated by phosphatidylinositol 3-kinase and extracellular signal-regulated kinase 1/2. Neoplasia. 2010;12(2):128-38. [PubMed ID: 20126471]. [PubMed Central ID: PMC2814351]. https://doi.org/10.1593/neo.91438.
-
8.
Sprowl JA, Reed K, Armstrong SR, Lanner C, Guo B, Kalatskaya I, et al. Alterations in tumor necrosis factor signaling pathways are associated with cytotoxicity and resistance to taxanes: A study in isogenic resistant tumor cells. Breast Cancer Res. 2012;14(1):R2. [PubMed ID: 22225778]. [PubMed Central ID: PMC3496117]. https://doi.org/10.1186/bcr3083.
-
9.
Wang H, Vo T, Hajar A, Li S, Chen X, Parissenti AM, et al. Multiple mechanisms underlying acquired resistance to taxanes in selected docetaxel-resistant MCF-7 breast cancer cells. BMC Cancer. 2014;14:37. [PubMed ID: 24447372]. [PubMed Central ID: PMC3900991]. https://doi.org/10.1186/1471-2407-14-37.
-
10.
Oh YK, Park TG. siRNA delivery systems for cancer treatment. Adv Drug Deliv Rev. 2009;61(10):850-62. [PubMed ID: 19422869]. https://doi.org/10.1016/j.addr.2009.04.018.
-
11.
Thiviyanathan V, Gorenstein DG. Aptamers and the next generation of diagnostic reagents. Proteomics Clin Appl. 2012;6(11-12):563-73. [PubMed ID: 23090891]. [PubMed Central ID: PMC4021847]. https://doi.org/10.1002/prca.201200042.
-
12.
Kufe DW. MUC1-C oncoprotein as a target in breast cancer: Activation of signaling pathways and therapeutic approaches. Oncogene. 2013;32(9):1073-81. [PubMed ID: 22580612]. [PubMed Central ID: PMC3621754]. https://doi.org/10.1038/onc.2012.158.
-
13.
Sayari E, Dinarvand M, Amini M, Azhdarzadeh M, Mollarazi E, Ghasemi Z, et al. MUC1 aptamer conjugated to chitosan nanoparticles, an efficient targeted carrier designed for anticancer SN38 delivery. Int J Pharm. 2014;473(1-2):304-15. [PubMed ID: 24905777]. https://doi.org/10.1016/j.ijpharm.2014.05.041.
-
14.
Ola MS, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis. Mol Cell Biochem. 2011;351(1-2):41-58. [PubMed ID: 21210296]. https://doi.org/10.1007/s11010-010-0709-x.
-
15.
Eder JP, Vande Woude GF, Boerner SA, LoRusso PM. Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin Cancer Res. 2009;15(7):2207-14. [PubMed ID: 19318488]. https://doi.org/10.1158/1078-0432.CCR-08-1306.
-
16.
Sun C, Sang M, Li S, Sun X, Yang C, Xi Y, et al. Hsa-miR-139-5p inhibits proliferation and causes apoptosis associated with down-regulation of c-Met. Oncotarget. 2015;6(37):39756-92. [PubMed ID: 26497851]. [PubMed Central ID: PMC4741860]. https://doi.org/10.18632/oncotarget.5476.
-
17.
Smith DM, Simon JK, Baker JJ. Applications of nanotechnology for immunology. Nat Rev Immunol. 2013;13(8):592-605. [PubMed ID: 23883969]. https://doi.org/10.1038/nri3488.
-
18.
Dinarvand M, Kiani M, Mirzazadeh F, Esmaeili A, Mirzaie Z, Soleimani M, et al. Oral delivery of nanoparticles containing anticancer SN38 and hSET1 antisense for dual therapy of colon cancer. Int J Biol Macromol. 2015;78:112-21. [PubMed ID: 25858880]. https://doi.org/10.1016/j.ijbiomac.2015.03.066.
-
19.
Liu Y, Yang Y, Ye YC, Shi QF, Chai K, Tashiro S, et al. Activation of ERK-p53 and ERK-mediated phosphorylation of Bcl-2 are involved in autophagic cell death induced by the c-Met inhibitor SU11274 in human lung cancer A549 cells. J Pharmacol Sci. 2012;118(4):423-32. [PubMed ID: 22466960]. https://doi.org/10.1254/jphs.11181fp.
-
20.
Uddin S, Hussain AR, Ahmed M, Al-Dayel F, Bu R, Bavi P, et al. Inhibition of c-MET is a potential therapeutic strategy for treatment of diffuse large B-cell lymphoma. Lab Invest. 2010;90(9):1346-56. [PubMed ID: 20531293]. https://doi.org/10.1038/labinvest.2010.108.
-
21.
You H, Ding W, Dang H, Jiang Y, Rountree CB. c-Met represents a potential therapeutic target for personalized treatment in hepatocellular carcinoma. Hepatology. 2011;54(3):879-89. [PubMed ID: 21618573]. [PubMed Central ID: PMC3181384]. https://doi.org/10.1002/hep.24450.
-
22.
Ganansia-Leymarie V, Bischoff P, Bergerat JP, Holl V. Signal transduction pathways of taxanes-induced apoptosis. Curr Med Chem Anticancer Agents. 2003;3(4):291-306. [PubMed ID: 12769774].
-
23.
Mhaidat NM, Wang Y, Kiejda KA, Zhang XD, Hersey P. Docetaxel-induced apoptosis in melanoma cells is dependent on activation of caspase-2. Mol Cancer Ther. 2007;6(2):752-61. [PubMed ID: 17308071]. https://doi.org/10.1158/1535-7163.MCT-06-0564.
-
24.
Creixell M, Peppas NA. Co-delivery of siRNA and therapeutic agents using nanocarriers to overcome cancer resistance. Nano Today. 2012;7(4):367-79. [PubMed ID: 26257819]. [PubMed Central ID: PMC4527553]. https://doi.org/10.1016/j.nantod.2012.06.013.
-
25.
Ye QF, Zhang YC, Peng XQ, Long Z, Ming YZ, He LY. Silencing Notch-1 induces apoptosis and increases the chemosensitivity of prostate cancer cells to docetaxel through Bcl-2 and Bax. Oncol Lett. 2012;3(4):879-84. [PubMed ID: 22741011]. [PubMed Central ID: PMC3362393]. https://doi.org/10.3892/ol.2012.572.
-
26.
Zheng C, Zheng M, Gong P, Deng J, Yi H, Zhang P, et al. Polypeptide cationic micelles mediated co-delivery of docetaxel and siRNA for synergistic tumor therapy. Biomaterials. 2013;34(13):3431-8. [PubMed ID: 23375952]. https://doi.org/10.1016/j.biomaterials.2013.01.053.
-
27.
Jun HT, Sun J, Rex K, Radinsky R, Kendall R, Coxon A, et al. AMG 102, a fully human anti-hepatocyte growth factor/scatter factor neutralizing antibody, enhances the efficacy of temozolomide or docetaxel in U-87 MG cells and xenografts. Clin Cancer Res. 2007;13(22 Pt 1):6735-42. [PubMed ID: 18006775]. https://doi.org/10.1158/1078-0432.CCR-06-2969.
-
28.
de Almeida CEB, Alves LN, Rocha HF, Cabral-Neto JB, Missailidis S. Aptamer delivery of siRNA, radiopharmaceutics and chemotherapy agents in cancer. Int J Pharm. 2017;525(2):334-42. [PubMed ID: 28373101]. https://doi.org/10.1016/j.ijpharm.2017.03.086.
-
29.
Cheng D, Cao N, Chen J, Yu X, Shuai X. Multifunctional nanocarrier mediated co-delivery of doxorubicin and siRNA for synergistic enhancement of glioma apoptosis in rat. Biomaterials. 2012;33(4):1170-9. [PubMed ID: 22061491]. https://doi.org/10.1016/j.biomaterials.2011.10.057.
-
30.
Kim C, Shah BP, Subramaniam P, Lee KB. Synergistic induction of apoptosis in brain cancer cells by targeted codelivery of siRNA and anticancer drugs. Mol Pharm. 2011;8(5):1955-61. [PubMed ID: 21793576]. [PubMed Central ID: PMC3185194]. https://doi.org/10.1021/mp100460h.
-
31.
Li X, Zhao Q, Qiu L. Smart ligand: Aptamer-mediated targeted delivery of chemotherapeutic drugs and siRNA for cancer therapy. J Control Release. 2013;171(2):152-62. [PubMed ID: 23777885]. https://doi.org/10.1016/j.jconrel.2013.06.006.