Abstract
Background:
Human ABCC3 (hABCC3) or multidrug resistance protein 3 (MRP3), third member of the subfamily C of the ABC proteins, is associated with multidrug resistance and treatment failure in acute leukemia. Hence, targeting this protein might be a useful approach to provide more effective drugs to overcome cancer drug resistance.Objectives:
The present study aimed at predicting the possible ligand binding sites (PBSs) on hABCC3 and examining the possible binding of different chemotherapeutic drugs to this protein.Methods:
To predict the PBSs on the hABCC3 transporter, a three-dimensional homology model of hABCC3 was generated in the current study based on bovine ABCC1 (bABCC1) using SWISS-MODEL and MODELLER programs, and then a molecular docking was qualified. Finally, binding affinities of 14 ligands including chemotherapeutic and immunosuppressive drugs, in addition to hABCC3 inhibitors, for hABCC3 were evaluated using binding free energies and the corresponding scores. Molecular docking was performed using AutoDock. Furthermore, Chimera, LigPlot, and Swiss PDB viewer (SPDBV) were used for interactive visualization and analysis of molecular structures.Results:
The two PBSs on hABCC3 were predicted using the blind docking method. Docking results in both PBSs showed that vincristine, doxorubicin, and daunorubicin had the highest binding affinities, respectively, with vincristine having the highest docking score.Conclusions:
In the current study, three drugs with the highest affinity for hABCC3 were introduced in order to take a step toward the possible hABCC3 targeting in drug-resistant acute leukemia. Furthermore, the application of in-silico methods in targeted cancer therapy, especially leukemia treatment, was highlighted.Keywords
hABCC3 MRP3 Acute Leukemia Homology Modeling Molecular Docking
1. Background
Multidrug resistance (MDR) is a major obstacle to successful chemotherapy, which leads to resistance to combination chemotherapy (1). One of the most important reasons for cancer drug resistance is to prevent the accumulation of anticancer drugs within the malignant cells. This general type of resistance is often caused by the overexpression of one of the oldest protein superfamilies called ATP (adenosine triphosphate)-binding cassette (ABC) transporters, which expel the drug molecules out of dividing tumor cells (2). Therefore, inhibition of the ABC transporters might be an effective approach to sensitize the resistant cells to chemotherapy drugs (3). MDR proteins (MRPs) are the transporters belonging to the “C” branch of the ABC superfamily. These family members transport and detoxify many chemically unrelated compounds and hamper the success of cancer pharmacotherapy (4). MRP3 or ABC subfamily C member 3 (ABCC3) is expressed in several tissue, predominantly in the kidney, liver, pancreas, and small intestine (5). ABCC3 gene is located on chromosome 17q22 and consists of 31 exons. It encodes 1527 amino acids, and the resulting 170- and 190-kDa membrane proteins have different glycosylation patterns. Human ABCC3 (hABCC3) is presumed to have three membranes and two large cytoplasmic domains. These two cytoplasmic domains contain nucleotide binding domains 1 and 2 (NBD-1 and -2) (6). The hABCC3 can confer the resistance against anticancer agents; a finding supported by previous studies showing the hABCC3 overexpression in multiple cancer types. Accordingly, it can be considered as a prognostic marker and therapeutic target (3). To explore the exact mechanism of action of an ABC transporter, its structure should precisely be explored. Despite efforts to determine the three-dimensional (3D) structure of ABC proteins, their high-resolution structure is still unknown since eukaryotic transporters are refracted in structural analysis (7).
Nowadays, in-silico models provide a fast, inexpensive, and non-laborious screening platform with promising and valuable results (8). In the current research, the homology modeling of hABCC3 was presented using the crystal structure of bovine ABCC1 (bABCC1) as a template. Furthermore, its docking to combination chemotherapeutic drugs was analyzed. Then, the results of the computational approaches were used to assess the binding characteristics of hABCC3 to the acute leukemia therapeutic agents such as etoposide, methotrexate, doxorubicin, vincristine, mitoxantrone, idarubicin, cytarabine, daunorubicin, prednisolone, and fludarabine, and four hABCC3 inhibitors including indomethacin, sulfinpyrazone, verapamil, and glyburide were discussed.
2. Methods
2.1. Software
Two programs including SWISS-MODEL server (https://swissmodel.expasy.org/) (9) and MODELLER 9.19 (10) were used to build homology models of hABCC3. Then, predicted models were evaluated by PROCHECK 3.5 (11) and the web-based version of ProSA (12). Molecular docking was performed by AutoDock 4.6.2 to combine chemotherapeutic agents (13). Finally, UCSF Chimera 1.11.2 (14), LigPlot 1.4.5 (15), and Swiss PDB viewer 4.10 (SPDBV) (16) were utilized for interactive visualization and molecular structure analysis.
2.2. Sequence Alignment and Homology Modeling
The FASTA sequence of hABCC3 (accession number: O15438) was retrieved from UniProt database (17) and used to find a suitable template with the most similarity to this transporter by basic local alignment search toolbox (BLAST) at PDB database (18). The bABCC1 (PDB ID: 5UJ9) (19) was an appropriate template for hABCC3. Then homology modeling of hABCC3 was performed by MODELLER 9.19 and SWISS-MODEL server.
2.3. Structure Analysis and Model Validation
The models of hABCC3 generated by SWISS-MODEL and MODELLER 9.19 were assessed by the Ramachandran plot (20) using PROCHECK, and checking the Z-score of structures obtained by ProSA (12). Finally, the model presented by MODELLER was selected for subsequent studies based on geometric and 3D alignment analyses. Then, the structures of the constructed hABCC3 model and the template were compared by Chimera.
2.4. Selection of the Ligand Molecules
First, in vitro hABCC3 substrates and inhibitors, as well as acute leukemia combination chemotherapeutic agents were obtained from DrugBank (21) (supplementary file Appendix 1). Then, ligand coordinates were obtained from DrugBank, PubChem Compound (22), and ZINC databases (23) (supplementary file Appendix 2).
2.5. Blind Docking
Molecular docking was performed using AutoDock 4.6.2 software to detect the ligand binding sites and probable binding pattern of abovementioned agents. In order to reduce the computational cost and time, a rigid docking protocol was considered through rigid receptor and rotatable ligands (13). AutoDockTools 1.5.6 was used to add the Gasteiger charges, merge non-polar hydrogen atoms, and define rotatable bonds (24). For blind docking, the two cytoplasmic domains (containing NBD-1 and NBD-2) were covered with two separate grid boxes of 126 × 126 × 126 grid points and 0.375 Å space between the points. The Lamarckian genetic algorithm was used to explore the best conformational space of the ligand (24). Accordingly, number of the runs in each docking experiment was set to 100, while other parameters were set to default. After collecting binding free energies of each ligand, the top results were selected. Then, the interaction between the protein and the ligands were precisely analyzed using LigPlot software.
3. Results and Discussion
3.1. Structure Prediction and Validation
Generally, a high sequence identity is preferred for high-quality models, although to establish a membrane transporter protein model of adequate quality, more than 30% similarity between the target and template sequences is required (25-27). Accordingly, blast of hABCC3 sequence (accession number: O15438) in the PDB database showed that the crystal structure of bABCC1 (PDB ID: 5UJ9) had the most query coverage and sequence identity (57%) among all other proteins (Figure 1). Hence, the 3D structure of bABCC1 was used as a template for model generation. The SWISS-MODEL and MODELLER 9.19 programs were used to build hABCC3 model (supplementary file Appendix 3). Then, the predicted model quality was evaluated by PROCHECK and ProSA programs. Further evaluation of the model by PROCHECK demonstrated that 99.3% of residues were in the allowed regions, showing the acceptable quality of the constructed model (supplementary file Appendix 4) (Table 1). In addition, Z-score obtained from ProSA tool was also used to evaluate the model quality. Generally, ProSA is used to assess the similarity rate between the input structure and the native proteins with the same size. Here, the Z-score for hABCC3 model was -12.7, which was within the acceptable range according to the X-ray and NMR studies (12) (supplementary file Appendix 5). These values supported the structural validity of the hABCC3 model.
PROCHECK Evaluation of hABCC3 Model
| Name | Most Favored Region, % | Additional Allowed Region, % | Generously Allowed Region, % | Disallowed Region, % |
|---|---|---|---|---|
| hABCC3 model | 90.3 | 8.1 | 0.9 | 0.7 |
Alignment between hABCC3 and bABCC1 (RMSD: 0.191), generated using Chimera

3.2. Blind Docking Analysis to Predict the Ligand Binding Sites
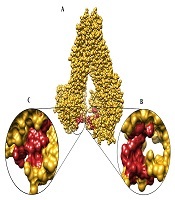
Blind docking is a reliable and useful approach to explore the possible ligand binding sites on a protein (28, 29). In fact, blind docking approach is used in a variety of docking studies to explore unknown binding sites on different proteins (7, 30-33). The blind docking approach was employed in the present study to dock the acute leukemia chemotherapeutic agents, considering the undefined ligand binding sites on hABCC3. These agents included etoposide, methotrexate, doxorubicin, vincristine, mitoxantrone, idarubicin, cytarabine, daunorubicin, prednisolone and fludarabine, as well as four hABCC3 inhibitors- i.e., indomethacin, sulfinpyrazone, verapamil, and glyburide. The current study employed two grid volumes covering two cytoplasmic domains of hABCC3 transporter. Then, the abovementioned ligands were blindly docked and the potent binding areas were predicted according to the most frequent binding sites and the highest binding affinities. In other words, the blind docking approach yielded 100 binding conformations for each ligand. The highest binding affinity of each compound belonged to a binding site that had the highest overlap among all ligands. Accordingly, all the agents examined in the study were bounded to two specific hotspot areas, located in two cytoplasmic domains of hABCC3 model; i.e., two ligand binding sites were exist. Structural analysis revealed that substantial amino acids involved in the conformation of PBS of cytoplasmic domain 1 (PBS-1) included Ser752, Gly753, Ser779, Ala780, Val781, Trp865, Thr866, Leu868, Glu869, Gly870, Ala871, Glu872, and Asp873, and amino acids involved in the conformation of PBS-2 were Leu877, Leu878, Ile879, Gln1371, Glu1451, Ala1452, Thr1453, Ala1454, Ala1455, Ile1456, Asp1457, and Leu1458. Moreover, residues such as Ser752, Gly753, Ser779, Ala780, and Val781 in PBS-1, and Gln1371, Glu1451, Ala1452, Thr1453, Ala1454, Ala1455, Ile1456, Asp1457, and Leu1458 in PBS-2 were conserved. These predicted binding sites were determined using AutoDockTools and Chimera (Figure 2) and then, the top binding free energy of each ligand in these sites was specified.
Two substrate binding sites in two cytoplasmic domains of the hABCC3 model; A, possible binding site of cytoplasmic domain 1 (PBS-1); B, PBS-2.

3.3. Docking Results of Cytoplasmic Domains 1 and 2
Docking energy of AutoDock showed the highest binding affinity to the PBS-1 in vincristine, followed by doxorubicin and daunorubicin. Interestingly, the three drugs with the highest binding affinities to PBS-2 were similar to those of PBS-1. In other words, these three drugs had more favorable interaction energies and higher affinities to both PBS-1 and PBS-2. The docking results of the ligands along with their binding energies for PBS-1 and PBS-2 are illustrated in Table 2. Residues involved in hydrogen binding and hydrophobic interactions in the binding modes are shown in Figures 3 and 4.
Docking Analysis of Ligands
| Ligand | AutoDock Docking Energy (kcal/mol) | ||
|---|---|---|---|
| PBS-1 | PBS-2 | ||
| 1 | Vincristine | -10.79 | -8.86 |
| 2 | Doxorubicin | -9.26 | -8.5 |
| 3 | Daunorubicin | -8.82 | -7.77 |
| 4 | Idarubicin | -8.66 | -7.1 |
| 5 | Mitoxantrone | -8.47 | -7.36 |
| 6 | Verapamil | -8 | -6.95 |
| 7 | Etoposide | -6.79 | -6.98 |
| 8 | Methotrexate | -6.64 | -6.3 |
| 9 | Glyburide | -6.5 | -5.67 |
| 10 | Prednisolone | -6.2 | -5.23 |
| 11 | Sulfinpyrazone | -5.83 | -6.35 |
| 12 | Fludarabine | -3.43 | -3.11 |
| 13 | Cytarabine | -3.34 | -3.28 |
| 14 | Indomethacin | Not seen | -6.03 |
Representations of the binding modes of the three drugs with more favorable interaction energy and stronger affinity to the PBS-1 of hABCC3 model (vincristine, doxorubicin, and daunorubicin, respectively), showing the ligands (purple), residues involved in hydrogen binding with the ligand (brown) along with their hydrogen bonds (green), and residues involved in non-bonded interactions (red spikes).

Representations of the binding modes of three drugs with highest binding affinities to the PBS-2 of hABCC3 model (vincristine, doxorubicin, and daunorubicin, respectively), showing the ligands (purple), residues involved in hydrogen binding with the ligand (brown) along with their hydrogen bonds (green), and residues involved in non-bonded interactions (red spikes).

In the present study, three drugs with the highest affinity for hABCC3 were introduced to take a step toward the possible hABCC3 targeting in drug-resistant acute leukemia. Furthermore, the application of in-silico methods in targeted cancer therapies, especially leukemia treatment, was highlighted in the current study.
References
-
1.
Wu Q, Yang Z, Nie Y, Shi Y, Fan D. Multi-drug resistance in cancer chemotherapeutics: Mechanisms and lab approaches. Cancer Lett. 2014;347(2):159-66. [PubMed ID: 24657660]. https://doi.org/10.1016/j.canlet.2014.03.013.
-
2.
Li W, Zhang H, Assaraf YG, Zhao K, Xu X, Xie J, et al. Overcoming ABC transporter-mediated multidrug resistance: Molecular mechanisms and novel therapeutic drug strategies. Drug Resist Updat. 2016;27:14-29. [PubMed ID: 27449595]. https://doi.org/10.1016/j.drup.2016.05.001.
-
3.
Balaji SA, Udupa N, Chamallamudi MR, Gupta V, Rangarajan A. Role of the drug transporter ABCC3 in breast cancer chemoresistance. PLoS One. 2016;11(5). e0155013. [PubMed ID: 27171227]. [PubMed Central ID: PMC4865144]. https://doi.org/10.1371/journal.pone.0155013.
-
4.
Chu XY, Huskey SE, Braun MP, Sarkadi B, Evans DC, Evers R. Transport of ethinylestradiol glucuronide and ethinylestradiol sulfate by the multidrug resistance proteins MRP1, MRP2, and MRP3. J Pharmacol Exp Ther. 2004;309(1):156-64. [PubMed ID: 14722317]. https://doi.org/10.1124/jpet.103.062091.
-
5.
Belinsky MG, Dawson PA, Shchaveleva I, Bain LJ, Wang R, Ling V, et al. Analysis of the in vivo functions of Mrp3. Mol Pharmacol. 2005;68(1):160-8. [PubMed ID: 15814571]. https://doi.org/10.1124/mol.104.010587.
-
6.
Fukushima-Uesaka H, Saito Y, Maekawa K, Hasegawa R, Suzuki K, Yanagawa T, et al. Genetic variations of the ABC transporter gene ABCC3 in a Japanese population. Drug Metab Pharmacokinet. 2007;22(2):129-35. [PubMed ID: 17495421]. https://doi.org/10.2133/dmpk.22.129.
-
7.
Hazai E, Bikadi Z. Homology modeling of breast cancer resistance protein (ABCG2). J Struct Biol. 2008;162(1):63-74. [PubMed ID: 18249138]. https://doi.org/10.1016/j.jsb.2007.12.001.
-
8.
Hattori H, Suminoe A, Wada M, Koga Y, Kohno K, Okamura J, et al. Regulatory polymorphisms of multidrug resistance 1 (MDR1) gene are associated with the development of childhood acute lymphoblastic leukemia. Leuk Res. 2007;31(12):1633-40. [PubMed ID: 17568669]. https://doi.org/10.1016/j.leukres.2007.04.009.
-
9.
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296-303. [PubMed ID: 29788355]. [PubMed Central ID: PMC6030848]. https://doi.org/10.1093/nar/gky427.
-
10.
Webb B, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics. 2014;47:561-32. [PubMed ID: 25199792]. https://doi.org/10.1002/0471250953.bi0506s47.
-
11.
Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: A program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26(2):283-91. https://doi.org/10.1107/s0021889892009944.
-
12.
Wiederstein M, Sippl MJ. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35(Web Server issue):W407-10. [PubMed ID: 17517781]. [PubMed Central ID: PMC1933241]. https://doi.org/10.1093/nar/gkm290.
-
13.
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785-91. [PubMed ID: 19399780]. [PubMed Central ID: PMC2760638]. https://doi.org/10.1002/jcc.21256.
-
14.
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605-12. [PubMed ID: 15264254]. https://doi.org/10.1002/jcc.20084.
-
15.
Laskowski RA, Swindells MB. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51(10):2778-86. [PubMed ID: 21919503]. https://doi.org/10.1021/ci200227u.
-
16.
Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714-23. [PubMed ID: 9504803]. https://doi.org/10.1002/elps.1150181505.
-
17.
The UniProt C. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158-69. [PubMed ID: 27899622]. [PubMed Central ID: PMC5210571]. https://doi.org/10.1093/nar/gkw1099.
-
18.
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235-42. [PubMed ID: 10592235]. [PubMed Central ID: PMC102472]. https://doi.org/10.1093/nar/28.1.235.
-
19.
Johnson ZL, Chen J. Structural basis of substrate recognition by the multidrug resistance protein MRP1. Cell. 2017;168(6):1075-1085 e9. [PubMed ID: 28238471]. https://doi.org/10.1016/j.cell.2017.01.041.
-
20.
Lovell SC, Davis IW, Arendall WB 3rd, de Bakker PI, Word JM, Prisant MG, et al. Structure validation by Calpha geometry: Phi,psi and Cbeta deviation. Proteins. 2003;50(3):437-50. [PubMed ID: 12557186]. https://doi.org/10.1002/prot.10286.
-
21.
Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A, et al. DrugBank 3.0: A comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011;39(Database issue):D1035-41. [PubMed ID: 21059682]. [PubMed Central ID: PMC3013709]. https://doi.org/10.1093/nar/gkq1126.
-
22.
Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202-13. [PubMed ID: 26400175]. [PubMed Central ID: PMC4702940]. https://doi.org/10.1093/nar/gkv951.
-
23.
Irwin JJ, Shoichet BK. ZINC--a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45(1):177-82. [PubMed ID: 15667143]. [PubMed Central ID: PMC1360656]. https://doi.org/10.1021/ci049714+.
-
24.
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19(14):1639-62. https://doi.org/10.1002/(sici)1096-987x(19981115)19:14<1639::aid-jcc10>3.0.co;2-b.
-
25.
Reddy Ch S, Vijayasarathy K, Srinivas E, Sastry GM, Sastry GN. Homology modeling of membrane proteins: A critical assessment. Comput Biol Chem. 2006;30(2):120-6. [PubMed ID: 16540373]. https://doi.org/10.1016/j.compbiolchem.2005.12.002.
-
26.
Gao C. Computational studies on membrane protein structures and protein-ligand binding affinities [dissertation]. University of Rochester; 2009.
-
27.
Forrest LR, Tang CL, Honig B. On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophys J. 2006;91(2):508-17. [PubMed ID: 16648166]. [PubMed Central ID: PMC1483079]. https://doi.org/10.1529/biophysj.106.082313.
-
28.
Hetenyi C, van der Spoel D. Efficient docking of peptides to proteins without prior knowledge of the binding site. Protein Sci. 2002;11(7):1729-37. [PubMed ID: 12070326]. [PubMed Central ID: PMC2373668]. https://doi.org/10.1110/ps.0202302.
-
29.
Hetenyi C, van der Spoel D. Blind docking of drug-sized compounds to proteins with up to a thousand residues. FEBS Lett. 2006;580(5):1447-50. [PubMed ID: 16460734]. https://doi.org/10.1016/j.febslet.2006.01.074.
-
30.
Espinoza-Fonseca LM, Trujillo-Ferrara JG. The existence of a second allosteric site on the M1 muscarinic acetylcholine receptor and its implications for drug design. Bioorg Med Chem Lett. 2006;16(5):1217-20. [PubMed ID: 16364641]. https://doi.org/10.1016/j.bmcl.2005.11.097.
-
31.
Gutierrez LJ, Enriz RD, Baldoni HA. Structural and thermodynamic characteristics of the exosite binding pocket on the human BACE1: A molecular modeling approach. J Phys Chem A. 2010;114(37):10261-9. [PubMed ID: 20806954]. https://doi.org/10.1021/jp104983a.
-
32.
Iorga B, Herlem D, Barre E, Guillou C. Acetylcholine nicotinic receptors: Finding the putative binding site of allosteric modulators using the "blind docking" approach. J Mol Model. 2006;12(3):366-72. [PubMed ID: 16372175]. https://doi.org/10.1007/s00894-005-0057-z.
-
33.
Mendez-Luna D, Martinez-Archundia M, Maroun RC, Ceballos-Reyes G, Fragoso-Vazquez MJ, Gonzalez-Juarez DE, et al. Deciphering the GPER/GPR30-agonist and antagonists interactions using molecular modeling studies, molecular dynamics, and docking simulations. J Biomol Struct Dyn. 2015;33(10):2161-72. [PubMed ID: 25587872]. https://doi.org/10.1080/07391102.2014.994102.