1. Background
Earthworms are the most common soil invertebrate found throughout the world in many ecosystems that helps agroecosystem sustainability (1). They play a prominent role in organic matter decomposition, modifying soil microbial dynamics, and nutrient composition, thereby being regarded as an ecosystem engineer (2). Earthworm gut is a tubular structure typically composed of mucus, organic amino acids, and minerals along with the presence of different kinds of microbial symbionts such as bacteria, protozoa, and micro-fungi (3). Gut of the earthworm is highly enriched with carbon and nitrogen with a ratio of seven (4), making it an ideal environment for bacteria to grow. Digestive enzymes like amylase, protease, cellulase, chitinase, lipase, and urease were reported in the digestive tract of earthworms (5). The intestines of earthworms contain a variety of microbes, enzymes, hormones, and other substances that aid in the rapid decomposition of soil and organic matter and the conversion of these materials into vermicompost (6). Microbial composition in the earthworm gut varies depending on the earthworm species, season, and routine feeding nature of the worms (7). The earthworms are categorized into three ecological groups based on their distinct feeding and burrowing habits, epigeic, endogeic, and anecic (8).
Perionyx excavatus (Perrier, 1872), the epigeic earthworm species are very small in size, having wonderful regenerative capability within a short period, and it has been widely used in the sustenance of soil fertility and breakdown of biological matter, which is facilitated by gut-associated microbiota (9). A few beneficial microorganisms in the animal gut microflora can also provide the host with vitamin B12 and riboflavin (10). Although it is known that the earthworm gut hosts a million microorganisms (11), little is known about their taxonomic positions at generic or species levels, and the taxonomic composition and functions of the microbial community are also unknown (12). The current study aimed to describe the fluorescence-emitting bacteria isolated from the gastrointestinal system of the earthworm, P. excavatus, in order to gain a better understanding of the microbe-earthworm symbiotic association.
2. Objectives
The present study was carried out to isolate fluorescence-emitting microbiota from the gut of earthworm, P. excavatus, and the fluorophores were partially characterized.
3. Methods
3.1. Culture and Maintenance of Earthworm, Perionyx excavatus
Earthworm, P. excavatus, was used in this study, collected from Tambaram, Chennai, and cultured in 'Regeneration and Stem cell Biology laboratory, International Research Centre (IRC) at Sathyabama Institute of Science and Technology, Chennai, Tamilnadu, India’. The earthworms were maintained in plastic tubs containing cow dung, organic wastes, soil, and leaf litters at 22 - 26°C room temperature. Six mature earthworms were measured and taken for the study, with a specific body mass of 150 - 300 mg, and were washed three times with distilled water to remove surface microbes from their skin.
3.2. Isolation of Earthworm Gut Microflora
The collected earthworms were again washed with 1x PBS solution for surface sterilization and then sacrificed by freezing. The gut was exposed using sterile dissecting pins, and the gut contents were carefully taken for the analysis, which were then serially diluted and spread plated on Luria Bertani (LB) agar plates (Hi-Media) under sterile environment, and kept overnight at 37°C. After incubation, bacterial colonies were observed using an ultra-violet (UV) transilluminator, and the fluorescence-emitting bacterial colonies were selected and sub-cultured.
3.3. Isolation and Purification of Fluorescence-Emitting Bacteria
Single colony of the fluorescence-emitting bacteria was isolated by quadrant streak method and transferred to the LB agar medium with the help of an inoculation loop. The plates were incubated at 37ºC for 24 hours. The obtained isolates were then subjected to morphological and molecular analysis for identifying the unknown bacterium. The isolated colonies were sub-cultured and stored in slants for further studies.
3.4. Identification of Fluorescence-Emitting Bacteria
To characterize the isolated fluorescence-emitting bacteria, initially, Gram staining method was carried out using Hi-Media reagents (Cat. No. K 001-1KT). The stained smear was observed under light microscope (100x oil immersion) and documented. In addition, the bacterial colonies were smeared on a clean glass slide, heat-fixed, and observed under fluorescence microscope (Euromex Upright fluorescence microscope), and the images were documented.
3.5. Molecular Identification
The bacterial strain was identified using the 16S ribosomal (rRNA) gene sequencing technique. To examine the 16S rRNA gene, genomic DNA from the bacterial strain was extracted (13), and the resulting DNA was examined by agarose gel electrophoresis, and the bands were documented. The 16S rRNA gene was amplified using universal forward and reverse primers (FP: 5'-AGAGTTTGATCCTGGCTCAG-3') (RP: 5'-TCTACGCATTTCACCGCTAC-3') and was then sent for sequencing, and the obtained sequences were analysed by using bioinformatics tools. The obtained nucleotide sequences were deposited to Gen Bank, and accession number was assigned as CP02665.1. Phylogenetic analysis was performed with nucleotide sequences using the maximum likelihood tree (ML) in molecular evolutionary genetics analysis (MEGA X) tool.
3.6. Thin-Layer Chromatography
Thin-layer chromatography (TLC) plate was coated with a thickness of 0.25 mm of layers slurry as stationary phase and hexane: Ethyl acetate as the mobile phase in a ratio of (9:1). The fluorescence-emitting bacterial colony was then centrifuged for five minutes at 10,000 rpm in LB broth to remove the supernatant. The sample was centrifuged again at 5,000 rpm for five minutes, and the pellets were dissolved in ethyl acetate and gently mixed. Now, with the help of capillary tube, the prepared samples were loaded on the silica, and the fluorescent bands were observed under ultra-violet (UV) trans-illuminator and documented accordingly.
3.7. UV-Vis Spectrophotometer
Structure elucidation of the isolated fluorescence bacterial isolate was performed using ultraviolet-visible (UV-Vis) spectrophotometer. The UV-visible absorption spectrum of the isolated fluorescence-emitting bacteria was tested using Shimadzu UV-2550 spectrophotometer at 320 - 800 nm. The OD values were observed, and a graph was plotted.
3.8. FTIR Analysis
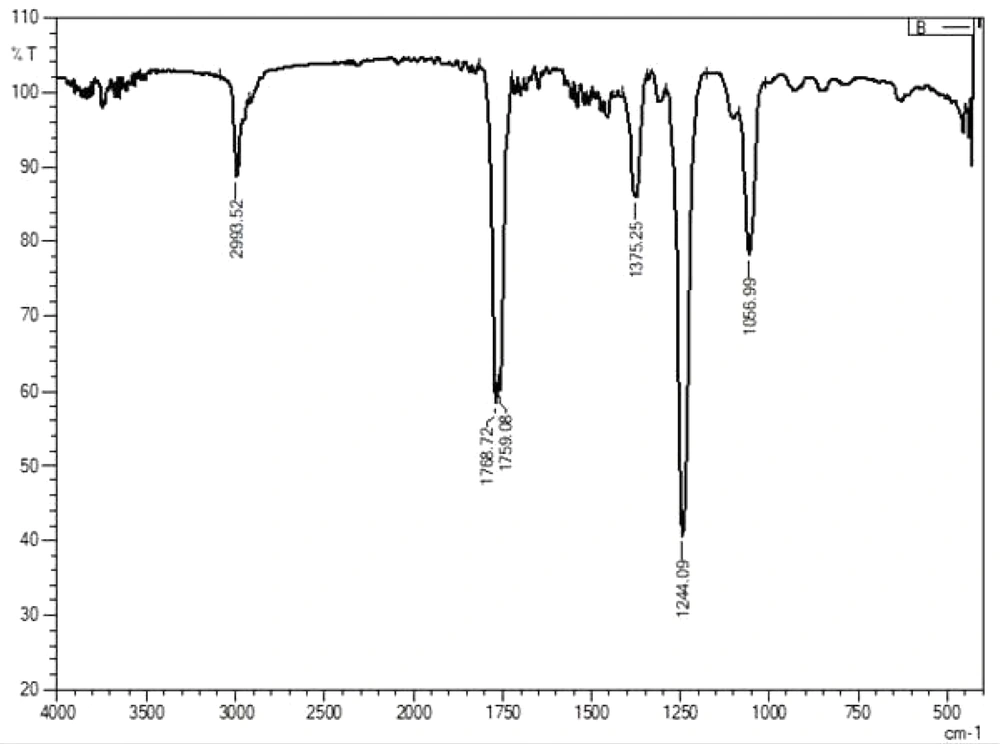
Fourier transform infrared spectroscopy (FTIR) was used to analyze the existence of specific chemical groups in the isolated fluorophores from the bacterial isolate. The vibrational spatial measurements were scored around 65 scans on a Digilab FTS 6,000 FT-IR spectrometer mounted with a Cesium Chloride detector. The analysis was performed in the spectral region of 400 to 4,000 cm-1 with a spectral resolution at 4 cm-1, and the background spectrum was obtained as a result using the computer software, ‘Origin’ from Perkin-Elmer spectrum.
3.9. Anti-inflammatory Assay
Anti-inflammatory activity was studied by inhibition of albumin denaturation, as per the protocol generated by Mizushima and Kobayashi, 1968. In this assay, samples were added to 0.5 ml of 1% bovine serum albumin (BSA) and incubated for 10 minutes at room temperature. After the incubation, it was heated to 51°C for 20 minutes. After the solution was cooled down to room temperature, absorbance at 660 nm was recorded, and the percent inhibition was calculated.
3.10. Regeneration Experiments
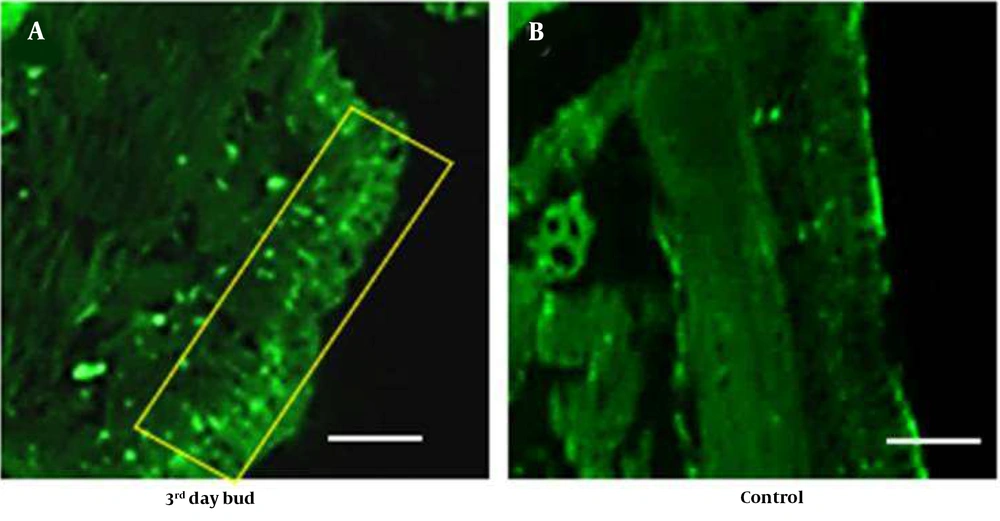
Two groups of P. excavatus earthworms were used in this study, each with six worms (control and test samples) and body masses ranging from 150 to 300 mg. Test groups will be amputated at the anterior junction of a 10 - 11th segment using a sterile surgical blade. The regeneration blastema (spongy transparent tissue) was observed on 3rd day after amputation, and the bud samples were collected for study. Another set, which serves as a control, was obtained by amputating the anterior segments 1 to 11. Both control and regenerating blastema samples (test groups) will be histologically examined to confirm their autofluorescence nature.
3.11. Tissue Section and Fluorescence Analysis
Autofluorescence property and tissue development patterns of earthworms during regeneration were studied using histology. Tissues were fixed with 10% formalin, cleared with xylene, and embedded in paraffin wax (Hi-Media). Thin sections (6 µm) were made using a microtome and examined under a fluorescence microscope.
3.12. Statistical Analysis
The mean and standard deviation for all data in this study was calculated using GraphPad Prism (GraphPad Software, USA). To obtain statistical significance, all other experiments are repeated at least three times. The results were considered statistically significant at P-value < 0.05.
4. Results
4.1. Isolation, Screening, and Identification of Fluorescence-Emitting Bacteria from Earthworm Gut
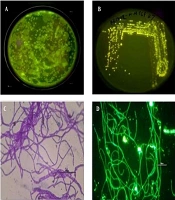
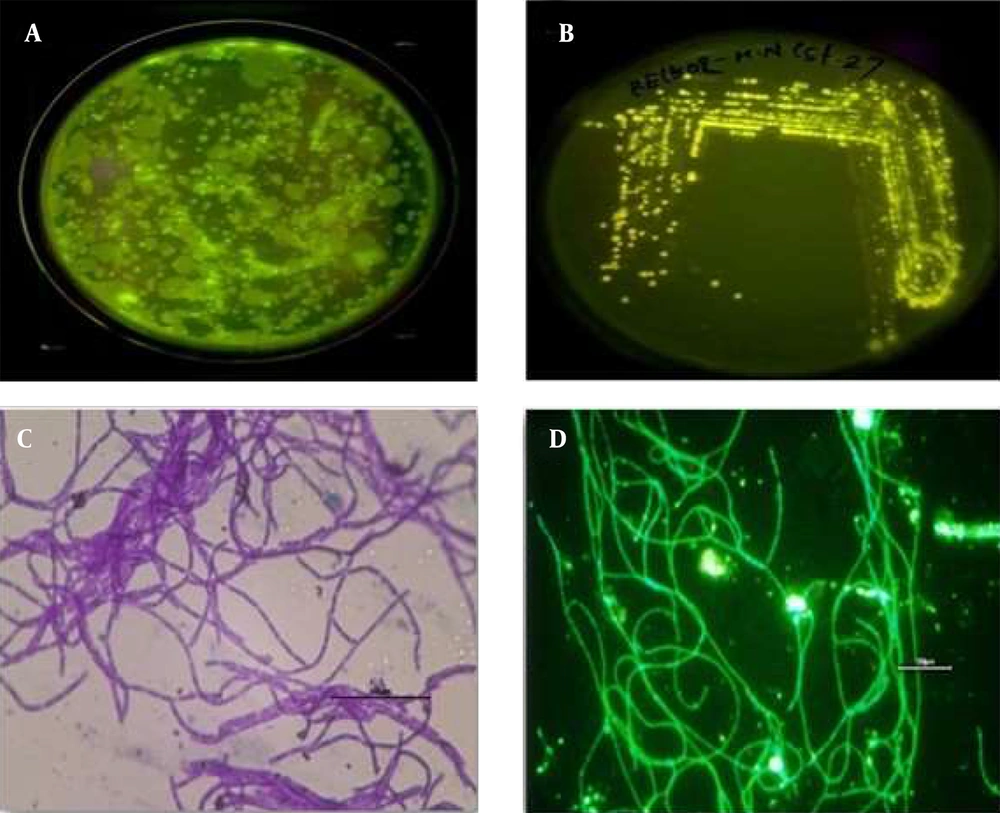
The gut samples of earthworm P. excavatus were serially diluted and plated in LB agar medium for the isolation of fluorescence-emitting bacterium. Interestingly, bright fluorescence-emitting bacterial colonies were observed on the Agar plates visualized under UV light (Figure 1A), and fluorescent-emitting colonies were picked and streaked separately on agar plate for getting pure colonies (Figure 1B). Sub-cultured bacterial colonies were used for Gram staining and subsequent experiments. Briefly, Gram staining revealed that the isolated unknown fluorescence-emitting bacterium was Gram-positive, and it is observed as rod-shaped bacterium having chain-like arrangements with smooth-edged colonies were observed under the light microscope (Figure 1C). Moreover, observation of heat-fixed unknown bacterium confirmed the presence of fluorophores inside the bacteria (Figure 1D), which emit green fluorescence.
4.2. 16S rRNA Amplification and Phylogenetic Analysis
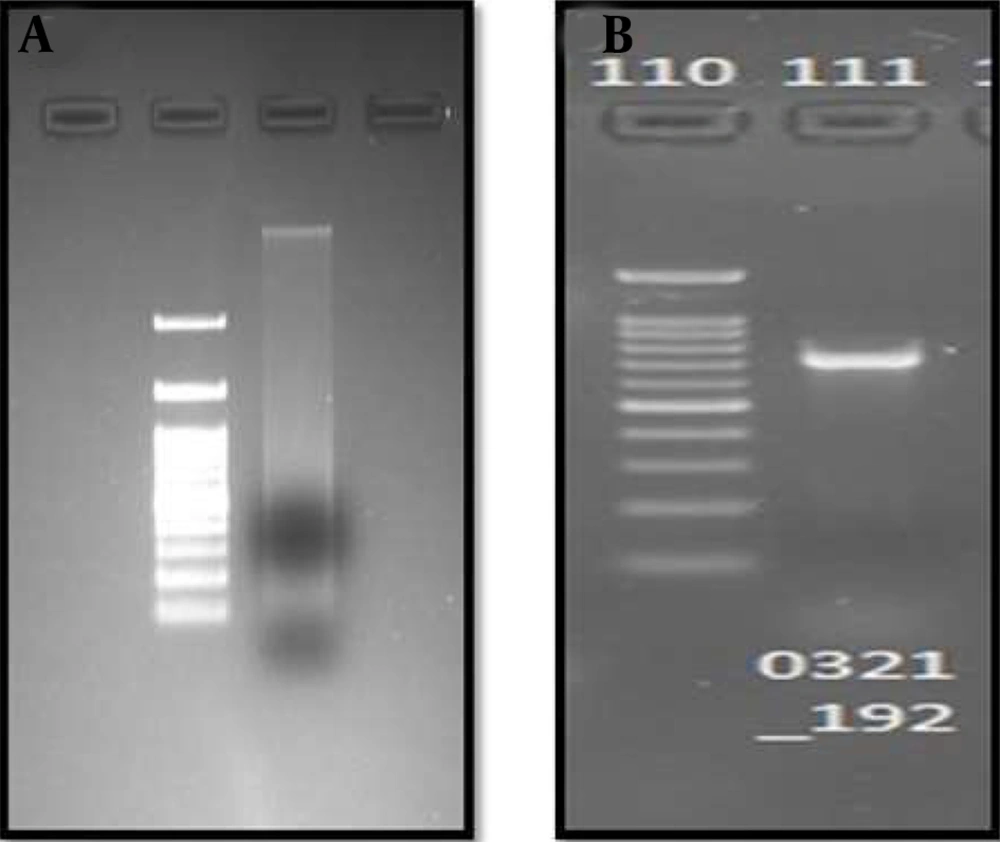
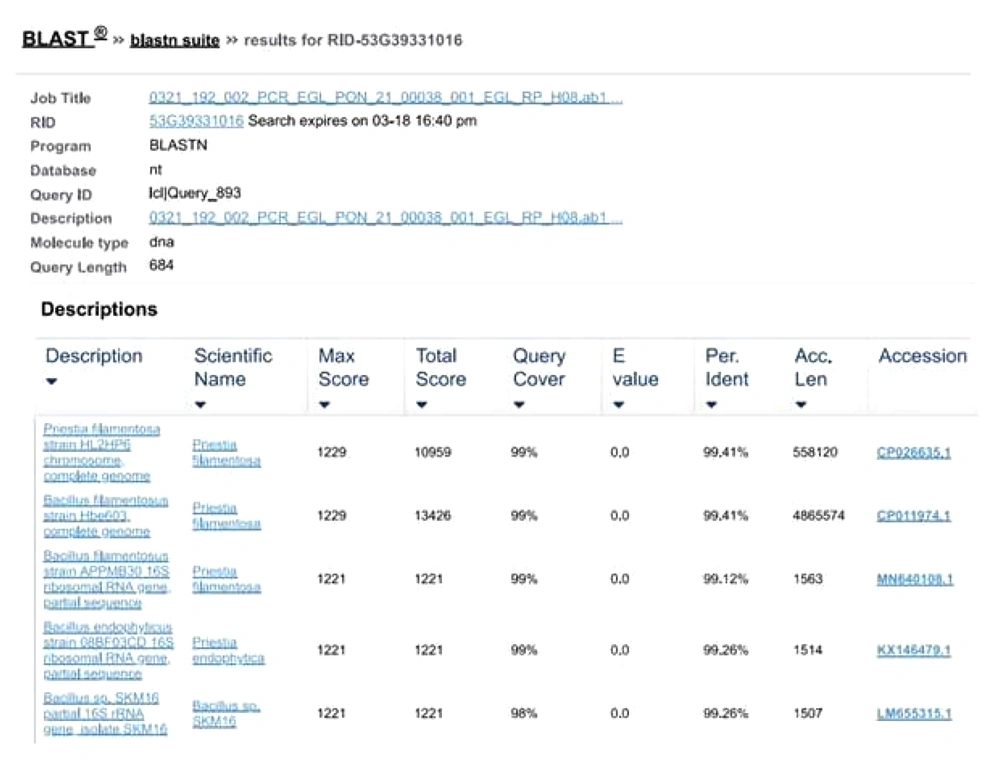
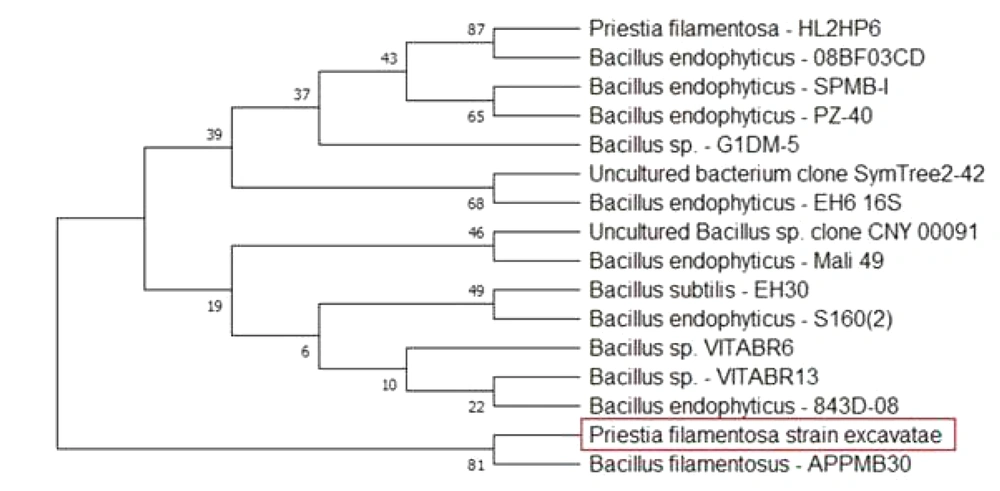
To perform the molecular characterization and identification of unknown fluorescence-emitting bacterium species, genomic DNA was extracted. DNA purified using a 0.8 percent agarose gel revealed sharp, high molecular weight bands of DNA (Figure 2A), and a PCR product of 684 bp was obtained, as shown in Figure 2B. The 16S rRNA gene was analyzed using the BLASTn (nucleotide BLAST) tool (Figure 3). The sequence of 16S rRNA gene of unknown bacterium showed 99% similarity with 16S rRNA gene of P. filamentosa. Phylogenetic tree was constructed based on the sequence comparison with closely related known bacterium species and the strain was confirmed as P. filamentosa, which was highlighted in the phylogenetic tree as shown in Figure 4.
4.3. Thin Layer Chromatography Analysis
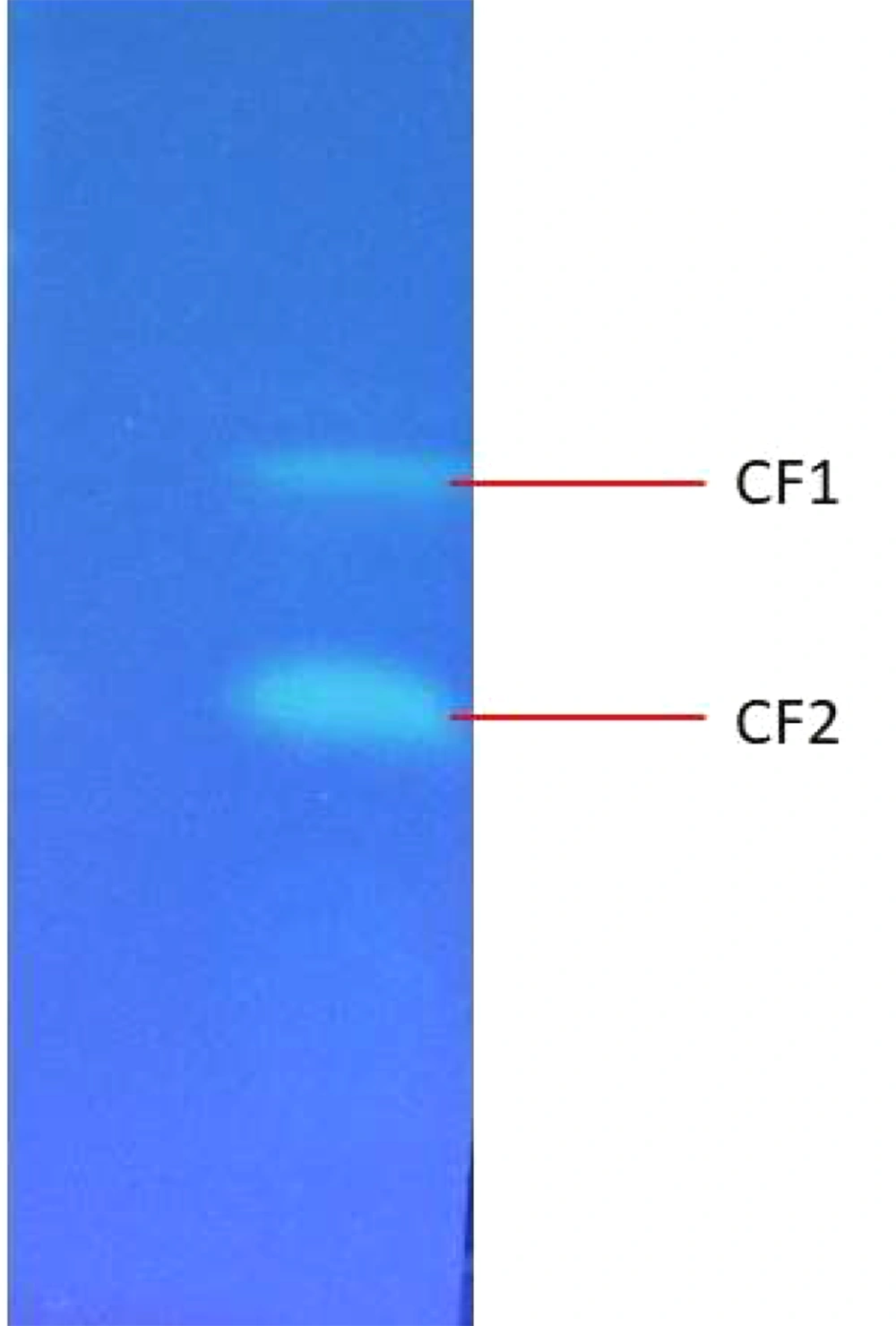
TLC results revealed that two fluorescent compounds are neatly visible under Ultra-UV trans-illuminator at 254-nm (Figure 5). The retardation factor (Rf) value for the unknown fluorescent compound 1 (CF1) and fluorescent compound 2 (CF2) was calculated as 2.2 and 4.0, respectively.
4.4. UV-Vis Spectrophotometer and FTIR Analysis of CF1 and CF2
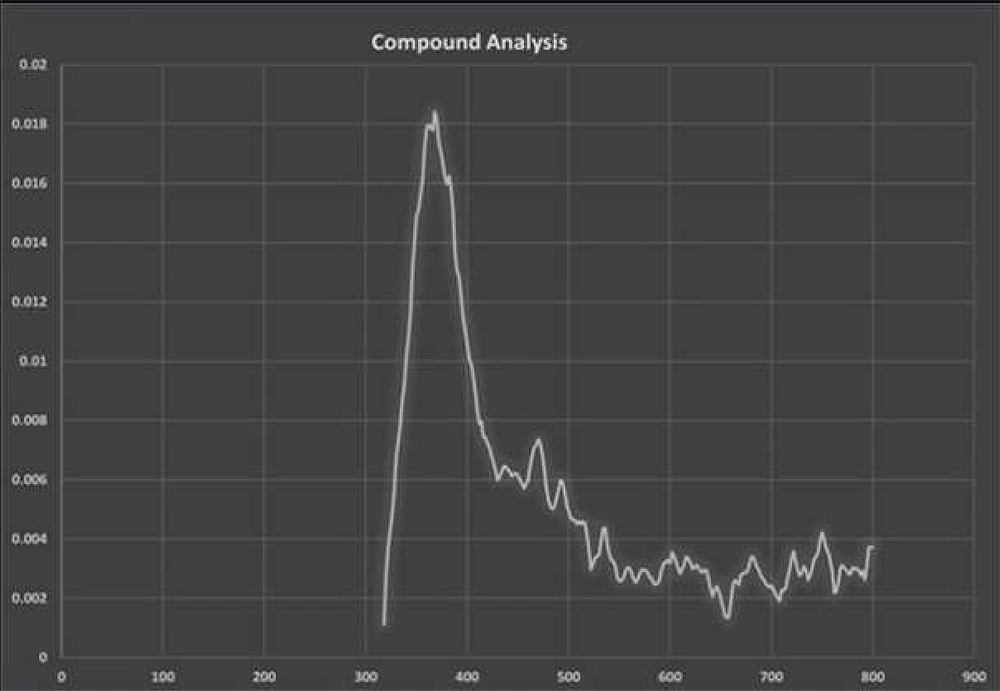
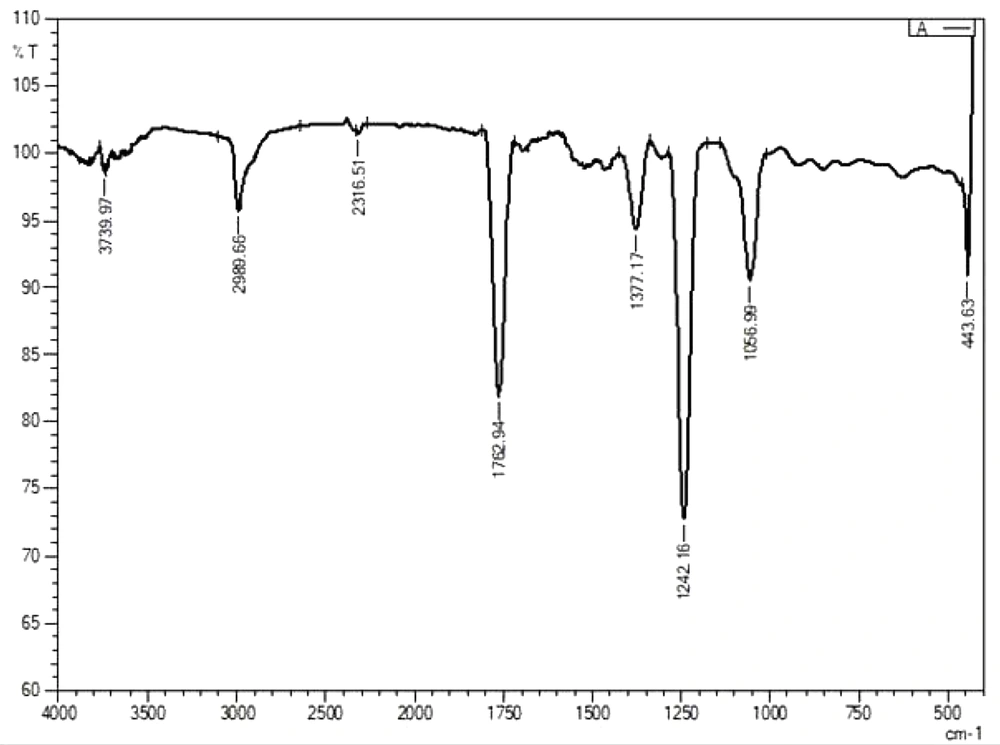
The fluorescence compounds, CF1 and CF2 were eluted from the TLC plate and then subjected to UV-spectroscopy analysis. Absorbance maxima were noted from 360 nm to 380 nm for CF1 and CF2, respectively. The experiment was precisely performed four times and consistent results were obtained, shown in Figure 6. FTIR data (Figures 7 and 8) clearly shows the functional groups of CF1 and CF2, which are more resembled with the riboflavin derivative. Further characterization is important to further confirm the data.
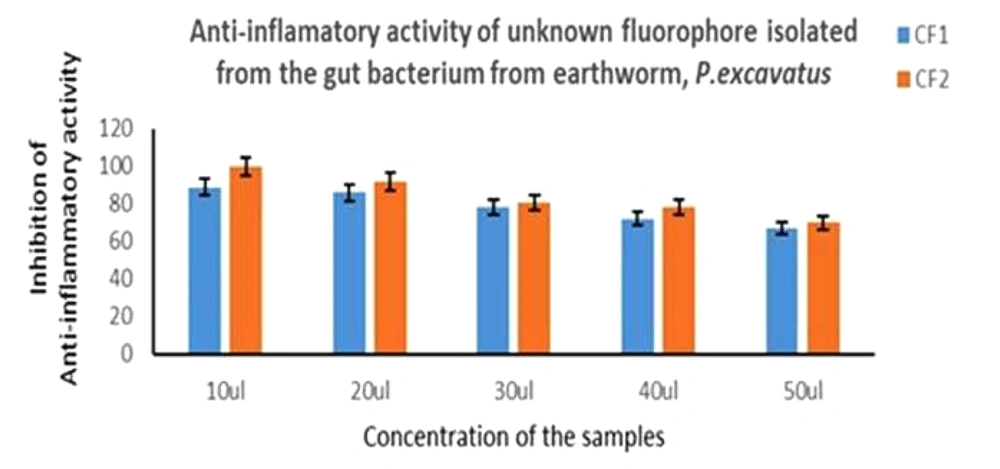
4.5. Anti-inflammatory Assay
Anti-inflammatory assay was performed with the fluorescence compounds, CF1 and CF2, which showed good activity. Briefly, based on the data, the protein degradation of CF1 was less compared to CF2, as shown in Figure 9.
4.6. Regeneration Studies and Autofluorescence
Earthworm, P. excavatus, has prodigious regeneration capability (14) has been studied extensively. Intense fluorescence was observed beneath the epithelial layer in the tissue sections of regenerating worms (Figure 10A). In control worms, a similar pattern was observed but compared to regenerating worms, fluorescence intensity was high in regenerating worm tissue sections compared to the control (Figure 10B).
5. Discussion
Riboflavin has been identified as a primary source of autofluorescence in coelomocytes of earthworm E. foetida (15). In our studies, the fluorophores produced in the earthworm skin epithelial layer are the same compared to the isolated fluorescence-emitting gut microbe, based on the UV spectrophotometry analysis. Based on the previous literature, UV-Vis absorbance and FTIR analysis, the fluorophores CF1 and CF2 might be riboflavin derivatives.
Several studies showed that Bacillus sp. was dominant in the gut of Eudrilus eugeniae (16), whereas Aeromonas hydrophila were the dominant culturable bacteria in E. foetida (17) and fluorescent pseudomonads in L. terrestris (18). Bacillus endophyticus, a bacteria found inside the gastrointestinal region of E. eugeniae, has been revealed to synthesize riboflavin, which assists in organ regeneration in amputated worms (19). However, our results show that P. filamentosa, a fluorescent bacterium, belongs to Bacillus family and possibly belongs to the new species, which may have a symbiotic association with P. excavatus regeneration.
Johnson Retnaraj Samuel et al. (20), reported the excitation and emission maxima of riboflavin are 450 and 524 nm, respectively. However, we observed absorbance peaks for CF1 and CF2 measured from 360 nm to 380 nm, respectively (Figure 6). Aromatic amino acids such as tyrosine, tryptophan, and phenylalanine exhibit distinct emission bands that range between 300 and 400 nm targets riboflavin (21). Thus, the obtained results revealed that isolated fluorescent compound might be a derivative of riboflavin.
The FTIR results were interpreted as shown in Figure 7 and 8, CF1 has recorded a peak wavelength of 3,739.97 cm-1 while CF2 have a peak wavelenght noted of 2,993.52 cm-1 indicates that it may be due to the presence of following functional groups such as N-H, O-H and C-H groups in the fluorophore. In addition, a wavelength shift was noted in CF1 from 2,343.51 cm-1 to 2,316.51 cm-1, indicating triple bond between C-C and/or C-N, respectively. Significantly, there is a huge peak noted at 1,768.72 cm-1 and 1,759.08 cm-1 inCF2 and single peak 1,762.94 cm-1 in CF2 indicates that has functional groups viz., C=C, C=O, and C=N. In addition, a shift at 1,375.25 cm-1 indicates methyl group is present in the CF1 and CF2. Moreover, a shift at the wavelength 1,242.16 cm-1 in CF1 and 1,244.09 in CF2 indicates the presence of C-N stretching amine groups (22). All of these functional groups identified by FTIR indicate that the fluorescence is mainly due to these reasons. Further studies such as proton and carbon NMR are important to fully characterize the fluorophores. Further studies are important to focus on mechanism of symbiotic association and the role of fluorophores in regeneration aspects.
5.1. Conclusions
Our study concludes that the isolated bacterium from earthworm, P. excavatus that has a fluorescence is identified as P. filamentosa. The partial structural characterization of the eluted unknown fluorophores, CF1 and CF2, confirms that it may be a riboflavin derivative. Anti-inflammatory assays confirm that the fluorophores have excellent anti-inflammatory properties. In addition, these fluorophores are expressed and noted in the epithelial cell layer of earthworm P. excavatus. Hence, these two fluorophores will be studied in-depth in the context of structural conformation, chemical composition, and functional characteristics for potential biotechnological applications.