Abstract
Background:
CCR5 is identified as one of the most important chemokine receptors with a major role in the creation of chemotaxis and mobilization of immunocompetent cells and moving them toward the liver for thorough cleaning of the virus. CCR5-59353 (C/T) is an important promoter polymorphism of chemokine receptor 5. Some studies showed a relationship between CCR5-59353 (C/T) polymorphism and clearance or persistence of hepatitis B virus (HBV) infection.Objectives:
The current study aimed at developing polymorphism CCR5-59353 (C/T) in Iranian patients with chronic HBV infection.Methods:
A total of 200 blood samples including 100 healthy controls and 100 HBsAg-positive patients were randomly selected. Samples were tested for HBsAg by the enzyme-linked immunosorbent assay (ELISA) and HBV-DNA by the polymerase chain reaction (PCR) method. Genomic DNA was extracted from blood buffy coat using the salting out method. CCR5-59353 (C/T) polymorphism was genotyped by the allele specific amplification (ASA) PCR. Chi-square test was used for statistical analysis.Results:
Five percent of control samples and 12% of patient samples had CC mutant genotype. Nevertheless, there was no significant difference in genotypes frequency of CCR5-59353 between the groups (P = 0.1).Conclusions:
It seems that CCR5-59353 polymorphism was not associated with chronic HBV infection outcome in the Iranian population. However, the frequency of CC genotype was higher in the patient group (12%) than the control group (5%).Keywords
1. Background
Hepatitis B virus (HBV) is a DNA virus belongs to Hepadnaviridae family. According to medical studies, it is considered as one of the most important infections. When it develops a chronic infection, it can lead to cirrhosis and liver cancer. HBV is responsible for 70% to 80% of chronic hepatitis cases in Iran; it is shown that HBV is the main cause of liver disease in Iran (1).
CCR5 is identified as one of the most important chemokine receptors with a major role in the creation of chemotaxis and mobilization of immunocompetent cells and moving them toward the liver for thorough cleaning of the virus (2). This receptor is particularly expressed on the surface of many immune cells such as granulocytes, macrophages, immature dendritic cells, lymphocytes CD8+ , and lymphocytes Th1 (3).
CCR5 gene is located on the short arm at position 21 of chromosome 3 (3p21.3) (4, 5).
The most important mutation is the 32 nucleotide deletion (CCR5-∆32) from the only exon of the gene, resulting in a shortened and deactivated CCR5 molecule. Homozygous genotype resulting from this mutation is highly resistant to HIV-1 infection and its heterozygous type is associated with 2 to 4 years of delay in the incidence of acquired immunodeficiency syndrome (AIDS). CCR5 gene has several single nucleotide polymorphism (SNP) in the promoter region reported to be associated with the progression of AIDS (6, 7). Single nucleotide polymorphisms existing in CCR5 gene promoter include positions -59029, -59353, and -59402 (8).
The reactions between chemokines and chemokine receptors in chronic viral hepatitis are important because when T-cells go to liver parenchyma, they become a tool to clear hepatitis virus from the liver cells. Hence, during viral hepatitis, chemokines can probably lead to the accumulation and activation of leukocytes in the tissues; consequently, chemokines reaction with their receptors may provide new targets to treat patients (9).
Recovery from illnesses caused by intracellular pathogens such as HBV is observed among people with a stronger and wider T-cell response (10); however, due to unknown reasons, the presence of CCR5 decreases the T-cells response (11). Thus, CCR5-∆32 mutation, which deactivates CCR5 molecules and decreases their presence at the cellular level, can lead to an increase in T-cell responses. Therefore, people with this mutation have a stronger response to HBV; for instance, Thio et al., found that CCR5-∆32 increased the chance of recovery from HBV infection and decreased the progression of chronic infection by about 50% (11).
So far, some studies showed that some polymorphisms in CCR5 gene are associated with changes in the level of mRNA transcription and regeneration, and can affect the outcomes of viral infections (12, 13). The current study investigated the effect of the CCR5-59353 polymorphism on chronic HBV infection. CCR5-59353 occurs in 1 of the 2 CCR5 gene promoter positions (8) and is the main factor responsible for the transcription of the gene (14, 15). Increasing the promoter activity can lead to increased transcription of CCR5 gene, thus increase of mRNA generation and enhancement of CCR5 expression on the surface of the cell.
2. Objectives
The current study aimed at evaluating polymorphism CCR5-59353 (C/T) in the Iranian patients with chronic HBV infection.
3. Methods
A total of 100 patients with chronic HBV infection and positive HBsAg test results for more than 6 months and negative HCV and HIV test results were enrolled in the study. In addition, a total of 100 blood samples taken from healthy individuals with negative HBsAg, HCV, and HIV test results referred to the clinical lab of Iranian blood transfusion organization (IBTO) were randomly selected. The current study was approved by the Ethics Committee of Higher Education Research Institute for transfusion medicine and all the participants signed a written informed consent form.
To conduct the study, 5 ml peripheral blood samples drawn in tubes containing potassium ethylenediaminetetraacetic acid (EDTA) anticoagulant were used; the samples were then centrifuged for 10 minutes at 2700 rpm and the buffy coat layer was isolated and used to extract genomic DNA.
The genomic DNA was extracted from buffy coat layer using a saturated salt solution (16). A spectrophotometer Nanodrop apparatus was used to measure the concentration and the ratio of light absorption at a wavelength of 260 to 280 nm. The samples with high levels of concentration and purity were used.
ASA-PCR (allele-specific amplification-polymerase chain reaction) was used to determine polymorphism genotypes (CCR5-59353) (C/T). Therefore, the primer sequences in Table 1 were used to determine polymorphism (17); the results showed that only the last allele at the end of the reverse primer 3′ was different among the 2 pairs of primers and their forward primers were similar.
Sequences of Primers Used for CCR5-59353(C/T) Polymorphism Genotypes
| Sequences (5′ to 3′) | Primer | Size of Amplicon (bp) | |
|---|---|---|---|
| 59029G-59353T | 59029G-59353C | ||
| Forward | GAGTGGAGAAAAAGGGGG | GAGTGGAGAAAAAGGGGG | 363 |
| Reverse | AGAATAGATCTCTGGTCTGAA<A> | AGAATAGATCTCTGGTCTGAA<G> | 363 |
Stages of the test were similar to a routine PCR, except that for every extracted DNA sample (template DNA) PCR was performed twice (each time with 1 of the primers). Thus, all the required steps were taken to carry out a PCR reaction in a final volume of 25 µL to determine CCR5-59353 polymorphism.
Reaction mix (25 µL) contained Master Mix 2X (TaKaRa, Japan) 12.5 µL, from each primer 0.5 µL (Iran, Sinacolon), 2.5 µL of DNA, and double distilled water. PCR was performed by Thermocycler instruments (Corbett) under the following conditions: an initial cycle of denaturation for 3 minutes at 98°C, followed by 35 cycles of denaturation at 95°C for 10 seconds, annealing at 55°C for 45 seconds, and extension for 1 minute at 72°C, followed by 72°C for 10 minutes. Electrophoresis was performed on agarose gel 1.5% for PCR product identification.
Data analysis was conducted with SPSS version 18 using Chi-square test. P ≤ 0.05 were considered significant.
4. Results
The mean age of the 200 participants in the current study (120 males and 80 females) was 40.56 ± 16.3 years. There was no significant difference between the controls and cases in terms of the mean age (40.12 vs. 40.99 years, P = 0.7).
For each sample, 2 PCR reactions were performed with specific primers. The results of electrophoresis of allele-specific PCR on agarose gel 1.5% helped to properly detect and determine the genotype of CCR5-59353 polymorphism. The PCR reaction conducted in both micro-tubes; it indicated heterozygous CT, but when PCR reaction was performed only in 1 micro-tube, it indicated homozygous CC or TT. The frequencies of CCR5-59353 polymorphism genotypes including genotypes TT, CT, and CC were 4%, 91%, and 5% in the control group and 4%, 84%, and 12% in the case group, respectively. Moreover, the frequency of allele C variant was 54% in the patients with chronic HBV infection and 50.5% in the healthy individuals (Table 2, Figure 1).
Allele Frequencies and Genotype Polymorphism of CCR5-59353 in the Patient and Control Groups
| Allele/Genotype | Frequency | Percentage of Frequency | ||
|---|---|---|---|---|
| Control Group | Patient Group | Control Group | Patient Group | |
| Allele | ||||
| T | 99 | 92 | %49.5 | %46 |
| C | 101 | 108 | %50.5 | %54 |
| Genotype | ||||
| TT | 4 | 4 | %4 | %4 |
| CT | 91 | 84 | %91 | %84 |
| CC | 5 | 12 | %5 | %12 |
CCR5-59353 Polymorphism Genotype Frequencies in the Controls and Patients Groups
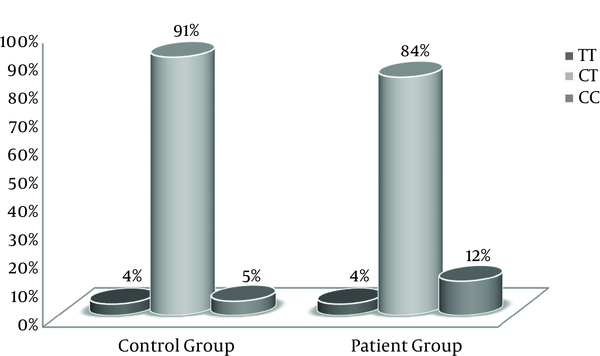
Statistical analysis was performed with SPSS version 18 using logistic regression. The frequency of allele C variant was 54% in patients with chronic HBV infection and 50.5% in healthy individuals with no statistically significant difference (OR = 1.15 P = 0.484). The frequency of CCR5-59353CC homozygote genotype was 12% in patients with HBV and 5% in healthy individuals with no significant difference between the 2 groups (OR = 2.49 P = 0.528) (Table 3).
Relationship of CCR5-59353 Polymorphism and Genotyping in the Patient with HBV and Control Groups
| Allele/Genotype | Control Group (n=100) | Patient Group (n=100) | P Value | OR | 95%CI |
|---|---|---|---|---|---|
| Allele | 0.484 | 1.15 | 0.78 - 1.70 | ||
| Genotype | |||||
| T/T | 4 | 4 | 0.471 | ||
| C/T | 91 | 84 | 0.772 | 0.73 | 0.8 - 6.36 |
| C/C | 5 | 12 | 0.528 | 2.49 | 0.15 - 42.42 |
5. Discussion
CCR5 acts as a receptor facilitating the entry of the HIV-1 virus into the host cell; several SNPs in the promoter region of the gene are associated with the progression of AIDS (6, 7). These SNPs are: position-59029 for nucleotide polymorphism of A to G, position-59353 for nucleotide polymorphism of C to T, and position -59402 for nucleotide polymorphism of A to G. Homozygous genotype of these SNPs is associated with accelerated progression of AIDS in patients with HIV infection (8, 18). According to Jang et al., pathogenesis of HIV mutant allele CCR5-59353C can be associated with HIV pathogenesis in a Korean population (19).
In another study, allele CCR5-59029A was identified as allele susceptible to generate HCV infection while allele CCR5-59029G was effective in preventing people from HCV infection (20). So far, no study was conducted to investigate the role of polymorphism -59353 (C/T) in HBV infection cases in Iran. Thus, the current study investigated the frequency of genotypic and allelic polymorphism in patients with HBV and healthy controls.
The current study did not aim to investigate the exact mechanism of the biological effect of the polymorphism CCR5-59353 C→T in the promoter region of CCR5 gene, but previous studies showed that some types of polymorphism make some changes in the level of CCR5 gene transcription and are associated with mRNA generation, which can affect the outcomes of HBV infection (12, 13). Studies showed that mutant alleles 59353C and 59029A can result in a 45% increase in the activity of the promoters, as compared with haplotype CCR5-59029G/CCR5-59353T (7). Increased promoter activity can lead to increased transcription of the CCR5 gene and increase mRNA generation, which consequently increases CCR5 expression on the cell surface.
In a study in South Korea conducted on patients with chronic HBV infection, the frequency of allele 59353C plus 59029A were significantly higher than that of patients with chronic HBV carriers. In other words, alleles CCR5 59029G/CCR5 59353T were spontaneously associated with the clearance of HBV (21).
Based on the collected data, it appears that CCR5 plays an important role in responding to HBV, because its expression is disrupted in natural killer (NK) cells, as well as cytotoxic T-cells in patients with long-term HBV infection. Additionally, due to the fact that the number of CCR5-positive NK cells and the intensity of CCR5 on NK cells of prolonged HBV infection are decreased during HBV infection (22), CCR5-59029 and -59353 polymorphisms may play important roles in the regulation of CCR5 expression.
In the current study, the frequency of CC genotype was higher in the patient group (12%) than the control group (5%).
There was no significant difference between the patients and controls in terms of homozygous mutant genotype (CC). In addition, no significant difference was observed between the 2 groups in terms of the frequency of mutant allele C. The results of the current study were consistent with those of a study by Kaur et al., conducted on 180 patients with chronic HIV-1 infection and 119 healthy individuals in the Northern regions of India. As the researchers reported, there was no significant difference between the 2 groups in terms of the frequency of SNP alleles in position -59353 in the CCR5 gene promoter, thus this type of polymorphism could not be effective in the incidence of HIV infection and progression of AIDS (23). However, for further studies on the role of polymorphism CCR5-59353 in chronic HBV infection, it is necessary to conduct population-based studies with larger sample sizes. Given the fact that Iran is a country with wide ethnic and genetic variations, the frequency of this type of polymorphism may vary among various ethnic groups.
Acknowledgements
References
-
1.
Alavian SM, Keyvani H, Rezai M, Ashayeri N, Sadeghi HM. Preliminary report of hepatitis B virus genotype prevalence in Iran. World J Gastroenterol. 2006;12(32):5211-3. [PubMed ID: 16937535].
-
2.
Ajuebor MN, Carey JA, Swain MG. CCR5 in T cell-mediated liver diseases: what's going on? J Immunol. 2006;177(4):2039-45. [PubMed ID: 16887960].
-
3.
Wong MM, Fish EN. Chemokines: attractive mediators of the immune response. Semin Immunol. 2003;15(1):5-14. [PubMed ID: 12495636].
-
4.
Hutter G, Neumann M, Nowak D, Kluter H, Hofmann WK. The beneficial effect of the ccr5-delta32 deletion in gvhd. Chemokine function or genecluster coexpression aside hla match? Vox Sang. 2010;99(1):495.
-
5.
Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722-5. [PubMed ID: 8751444]. https://doi.org/10.1038/382722a0.
-
6.
Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, et al. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381(6584):667-73. [PubMed ID: 8649512]. https://doi.org/10.1038/381667a0.
-
7.
McDermott DH, Zimmerman PA, Guignard F, Kleeberger CA, Leitman SF, Murphy PM. CCR5 promoter polymorphism and HIV-1 disease progression. Multicenter AIDS Cohort Study (MACS). Lancet. 1998;352(9131):866-70. [PubMed ID: 9742978].
-
8.
Mummidi S, Ahuja SS, Gonzalez E, Anderson SA, Santiago EN, Stephan KT, et al. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med. 1998;4(7):786-93. [PubMed ID: 9662369].
-
9.
Berger EA, Murphy PM, Farber JM. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657-700. [PubMed ID: 10358771]. https://doi.org/10.1146/annurev.immunol.17.1.657.
-
10.
Rehermann B, Lau D, Hoofnagle JH, Chisari FV. Cytotoxic T lymphocyte responsiveness after resolution of chronic hepatitis B virus infection. J Clin Invest. 1996;97(7):1655-65. [PubMed ID: 8601631]. https://doi.org/10.1172/JCI118592.
-
11.
Thio CL, Astemborski J, Bashirova A, Mosbruger T, Greer S, Witt MD, et al. Genetic protection against hepatitis B virus conferred by CCR5Delta32: Evidence that CCR5 contributes to viral persistence. J Virol. 2007;81(2):441-5. [PubMed ID: 17079285]. https://doi.org/10.1128/JVI.01897-06.
-
12.
Wu L, Paxton WA, Kassam N, Ruffing N, Rottman JB, Sullivan N, et al. CCR5 levels and expression pattern correlate with infectability by macrophage-tropic HIV-1, in vitro. J Exp Med. 1997;185(9):1681-91. [PubMed ID: 9151905].
-
13.
Paxton WA, Liu R, Kang S, Wu L, Gingeras TR, Landau NR, et al. Reduced HIV-1 infectability of CD4+ lymphocytes from exposed-uninfected individuals: association with low expression of CCR5 and high production of beta-chemokines. Virology. 1998;244(1):66-73. [PubMed ID: 9581779]. https://doi.org/10.1006/viro.1998.9082.
-
14.
Liu R, Zhao X, Gurney TA, Landau NR. Functional analysis of the proximal CCR5 promoter. AIDS Res Hum Retroviruses. 1998;14(17):1509-19. [PubMed ID: 9840284]. https://doi.org/10.1089/aid.1998.14.1509.
-
15.
Moriuchi H, Moriuchi M, Fauci AS. Cloning and analysis of the promoter region of CCR5, a coreceptor for HIV-1 entry. J Immunol. 1997;159(11):5441-9. [PubMed ID: 9548484].
-
16.
Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. [PubMed ID: 3344216].
-
17.
Tang J, Rivers C, Karita E, Costello C, Allen S, Fultz PN, et al. Allelic variants of human beta-chemokine receptor 5 (CCR5) promoter: evolutionary relationships and predictable associations with HIV-1 disease progression. Genes Immun. 1999;1(1):20-7. [PubMed ID: 11197301]. https://doi.org/10.1038/sj.gene.6363640.
-
18.
Clegg AO, Ashton LJ, Biti RA, Badhwar P, Williamson P, Kaldor JM, et al. CCR5 promoter polymorphisms, CCR5 59029A and CCR5 59353C, are under represented in HIV-1-infected long-term non-progressors. The Australian Long-Term Non-Progressor Study Group. AIDS. 2000;14(2):103-8. [PubMed ID: 10708279].
-
19.
Jang DH, Choi BS, Kim SS. The effects of RANTES/CCR5 promoter polymorphisms on HIV disease progression in HIV-infected Koreans. Int J Immunogenet. 2008;35(2):101-5. [PubMed ID: 18218038]. https://doi.org/10.1111/j.1744-313X.2007.00743.x.
-
20.
Al-Sharif FM, Al-Jiffii O, Azhar EI. Ccr5 a32 and ccr5-59029 allele frequency among hepatitis c virus infected and in non-hcv infected saudi population. World Appl Sci J. 2011;13(3):611-4.
-
21.
Ahn SH, Kim DY, Chang HY, Hong SP, Shin JS, Kim YS, et al. Association of genetic variations in CCR5 and its ligand, RANTES with clearance of hepatitis B virus in Korea. J Med Virol. 2006;78(12):1564-71. [PubMed ID: 17063508]. https://doi.org/10.1002/jmv.20739.
-
22.
Arababadi MK, Pourfathollah AA, Jafarzadeh A, Hassanshahi G. Decreased expression of CCR5 on the NK cells in occult HBV infected patients. Lab Med. 2010;41(12):735-8.
-
23.
Kaur G, Singh P, Rapthap CC, Kumar N, Vajpayee M, Sharma SK, et al. Polymorphism in the CCR5 gene promoter and HIV-1 infection in North Indians. Hum Immunol. 2007;68(5):454-61. [PubMed ID: 17462514]. https://doi.org/10.1016/j.humimm.2007.01.016.