Abstract
Keywords
Genome Editing Chimeric Antigen Receptor CRISPR TALEN ZFN T-Cell Receptor Cancer Immunotherapy
1. Context
T cells are an important component of the immune system, with hundreds of billions of them present in the lymph tissues and circulatory system. T cells can kill diseased (tumor) cells. Detection and response to diseased cells are performed by the interaction of T-cell receptors (TCRs) with the antigen presented by the major histocompatibility complex (MHC) of tumor cells. In a limited range of cancers, including melanoma and virus-associated malignancies, tumor-specific T cells are stimulated and account for tumor regression. In melanoma, these tumor-specific T cells can often be isolated from tumor-infiltrating lymphocytes and then re-injected into the patient after activation and proliferation in the laboratory (1). However, in most cancers, tumor cells pose significant challenges to deactivate T cells.
Tumors evolve using strategies, such as evading or suppressing anti-tumor immune response; accordingly, tumor cells blind T cells to their presence by the reduction of antigen processing or MHC expression. In addition, tumor cells create an environment suppressing activity and reducing the survival and migration of T cells. Furthermore, growth factors and immunosuppressive compounds may be simultaneously secreted from macrophages and granulocytes, promoting tumor growth and reducing immune activity. In this situation, the stimulation of endogenous T cells against the tumor is often ineffective (2).
However, these challenges can be overcome by the genetic modification of T cells, reprogramming them by inserting genes that provide specificity against tumor antigens to detect and destroy cancer cells (3). To date, adoptive T-cell therapy has shown promising potential for the treatment of cancer patients. Recent clinical trials involving treatment with genetically engineered T cells have shown significant efficacy. It is now expected that immunotherapy with T cells will not only control tumor progression but also may even treat cancer in some patients. In contrast, in clinical studies, despite the well-established efficacy of the therapy, severe side effects have been reported, suggesting that control and safety assessment are essential for the development of this treatment. There are new approaches to T cell-based immunotherapy, including reducing side effects, targeting new antigens, and using allogeneic cells to broaden the immunotherapy horizon into less harmful and effective cancer treatment (4).
Successful reports of treating cancer patients with T-cell-based therapies have encouraged academics and industry to develop such treatments for clinical use. However, it is becoming increasingly clear that this approach is auspicious but not without significant consequences. In particular, the use of synthetically engineered TCRs requires extreme caution, as these TCRs are never checked by the thymus system, and T-cells with an autoreactivity response are not separated and may react with normal tissues (5). Moreover, the patient-specific nature of these therapies implies that the T-cell product should be made for each patient. This patient-specific manufacturing method increases therapy costs and hinders product quality given the low number and reduced activity of T cells of the patients who have undergone extensive chemotherapies. These difficulties can theoretically be overcome using allogeneic T cells.
However, to obtain a safe allogeneic T-cell product, two problems should be addressed, including the possible rejection of the allogeneic chimeric antigen receptor (CAR) T cells and the cytotoxic effect of alloreactive T cells within the T-cell product. Advances in genome engineering provided powerful tools to overcome these challenges by the genetic knockout of the genes responsible for these undesired characteristics of the allogeneic T cells. This review discusses the challenges in allogeneic T cell therapies and the application of genome-editing technology to address these challenges.
2. Immunotherapy with Engineered T Cells
Although tumor cells are derived from healthy tissues, they may express specific molecules called tumor-associated antigens, which are the hallmarks of tumor cells’ differentiation from normal tissues. Tumor-associated antigens can be surface proteins expressed on the cell surface or presented as peptide fragments on the MHC molecules (6). T cells are rarely able to respond to tumor-associated antigens through their TCRs because tumors are derived from normal tissue, and the immune tolerance process that naturally occurs when T cells recognizing normal cells prevents them from responding to the tumor antigens. In the process of inducing immune tolerance occurring throughout an individuals’ life, tumor-specific T cells are either removed or become non-reactive to this type of tumor antigens as they are not reactive to the self-antigens (1).
However, genetically engineered T cells can be produced responding to tumors. These cells are produced by transducing genes encoding cell surface receptors that are capable of detecting tumor-associated antigens. T cells modified with these receptors could detect tumor antigens and kill tumor cells. T-cell engineering basics for cancer immunotherapy produce tumor-targeted T cells to overcome the obstacles that inhibit the induction and implementation of effective immune responses (7). Two categories of antigen, namely receptors physiological TCRs and synthetic CAR molecules, are used to reprogram T-cell specificity against tumor cells.
2.1. TCR-Engineered T Cells
TCRs are isolated from a clone of T cells with a high affinity for the target tumor antigen. These clones can be isolated from patients with very reactive but insignificant T-cell clones, targeting and destroying tumor cells (8). Nonetheless, the difficulty of isolation of these clones and their potentially low affinity for target antigens limit their application. The TCR can also be harvested from animal model T-cell clones (9).
2.2. Chimeric Antigen Receptor T Cells
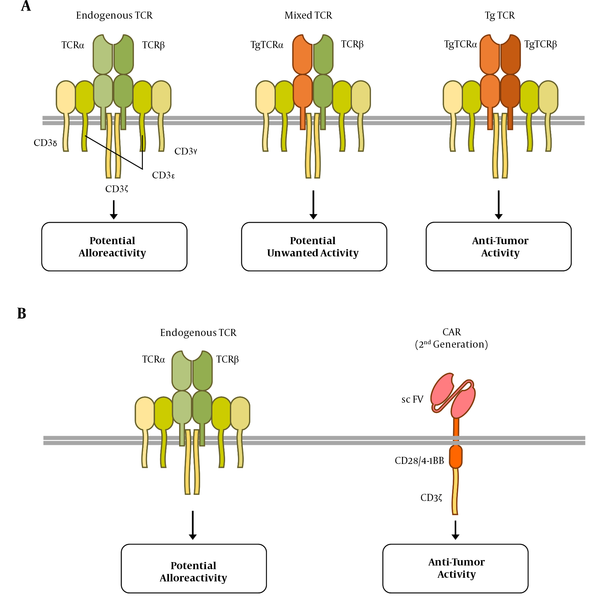
One type of genetically engineered T cell-based therapies is CAR-T-cell therapy, which has been reported to be remarkably effective in the treatment of B-cell malignancies. These therapies entail the injections of T cells which have been genetically modified to express a CAR protein. The synthetic CAR construct contains a single-chain variable fragment binding to the tumor surface antigen, transmembrane domain, and an intracellular signal transduction portion derived from CD3z and co-stimulatory molecules, such as CD28 or 4-1BB (5). The CAR detects intact antigens in a non-MHC-restricted manner and can be used in all individuals regardless of their human leukocyte antigen (HLA) type, which is an advantage of CAR over TCR-engineered cells (3, 10).
The cytoplasmic signaling domain is also essential for CAR design, derived from molecules that initiate lymphocyte signals. The CAR interaction with tumor-associated antigens induces associated molecules, phosphorylation of signaling domains, activation of downstream kinase cascades, gene expression, and finally activation of CAR T cells and their response to diseased cells. The signal-initiating molecules used in the CAR design include the ζ-chain of the TCR-CD3 complex and the γ-chain, the high-affinity receptor for immunoglobulin E (FcεRI). Since this single extracellular chain is derived from antibodies specific for tumor-associated antigens, CAR will be MHC-independent (10). In recent decades, the design of physiological TCRs and CARs has been continuously improved.
3. Challenges of Immunotherapy with Engineered Autologous T Cells
The genetic modification of autologous peripheral blood T lymphocytes for the production of targeted tumor T cells is an approach that has been well developed in several scientific centers. The potency of CAR and TCR therapies using this process has been well demonstrated by the clinical outcome from the New York esophageal squamous cell carcinoma (NY-ESO-1) TCR and CD19 CAR T cell (7). In this procedure, the patient’s T cells are harvested and transferred to a good manufacturing practice. T cells meeting acceptable criteria are genetically engineered to express either a new TCR or a CAR on their surface and re-injected to the patient after a short period of in vitro proliferation.
Most of the recent clinical trials based on TCR and CAR T cells use autologous engineered T cells; however, the low quality and quantity of these T cells may hamper the manufacturing process in these methods (11). Furthermore, the custom-made manufacturing of engineered T cells, which should be produced for each patient from their own T cells (12), leads to high production costs and complexity of manufacturing genetically modified T cells, which ultimately limits the production and use of such therapies in specialized centers (13). The inherent delays in a therapeutic product’s manufacturing process will also cause severe adverse outcomes for patients. It will also be impossible to produce custom-made manufacturing for patients with lymphopenia due to previous chemotherapy (14).
4. Immunotherapy with Allogeneic Off-the-Shelf Engineered T Cells
If high potential tissue-adaptive T cells are readily available, the promising results of engineered T cell-based therapies can be developed. The autologous approach is a proven one; however, custom-made manufacturing poses challenges in some cases. Using allogeneic T cells could provide an opportunity to overcome the limitation of autologous therapies. Although T cells can be easily harvested from donors in this approach, their application has a high potential for alloreactivity (15). An off-the-shelf T-cell product is a universal product produced from healthy allogeneic donors and can be transfused to any patient without causing graft-versus-host disease (GVHD) or rejecting the transferred T cells (16). For the generation of an off-the-shelf T-cell product, it is necessary to use methods that (1) increase histocompatibility between donor and recipient cells and prevent the rejection of T cells by the host; and (2) inhibit the alloreactivity of the product so that the adoptively transferred T cells do not exert cytotoxicity against recipients’ cells.
The expression of MHCs on the surface of allogeneic T cells causes their immediate rejection by the host immune system (11). The MHC class I and class II proteins play an important role in initiating the adaptive immune response. Both classes presented various peptides recognized by T cells. The MHC I presented peptides on most of the nucleated cells, which were recognized by cytotoxic CD8+ T cells; however, class II presented peptides on only antigen-presenting cells, such as dendritic cells, macrophages, or B cell, recognized by CD4+ T cells, leading to activation and differentiation from effector cells.
For the first time in 1945, the immune-mediated rejection of tissue allografts was discovered by Peter Medawar. The MHC was described as one of the most polymorphic genes in eukaryotes and the main barrier to successful tissue graft (17). T cells recognize non-self-allogeneic MHC molecules, resulting in activation and tissue rejection known as alloreactivity. Alloreactivity could be direct or indirect. Direct alloreactivity refers to intact mismatched MHC expressed on allogeneic cells and could be recognized with both naïve or memory T cells (18). Indirect alloreactivity refers to polymorphic peptides derived from allogeneic MHC molecules and presented in the antigen-binding groove of self-MHC molecules (19). Therefore, the prevention of MHC expression is considered a potential way to avoid the allogeneic rejection of the transferred T Cells Using Different Strategies, Including Genome Editing.
The endogenous TCRs of allogeneic T cells may recognize the recipient’s antigens as alloantigen, leading to GVHD (Figure 1). In the TCR-engineered T cells, the assembly of endogenous TCR alpha or beta chains with their transgenic counterparts may lead to the formation of mixed TCR complexes (Figure 1A), which might be self- or allo-reactive (20). One strategy to overcome these challenges is the elimination of endogenous TCR expression, which leaves the control of T cell activation and proliferation to the transgenic TCR or CAR. This is reachable through the revolutionary genome editing technologies discussed in the next sections.
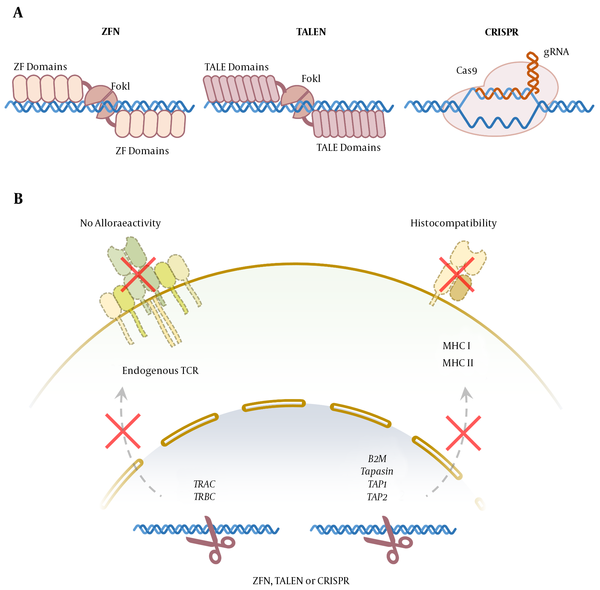
5. Genome-Editing Technologies
The induction of a double-strand break (DSB) by engineered nucleases leads to the induction of damage response pathways to endogenous deoxyribonucleic acid (DNA). The DNA damage repair is performed by homology-directed repair in the presence of an appropriate DNA pattern. In the absence of a template DNA, the dominant repair pathway is error-prone nonhomologous end joining, causing the insertion or deletion of few nucleotides in the DSB point and finally losing gene function (21). This study briefly describes genome editing technologies that have been used in the context of T-cell engineering.
5.1. Zinc Finger Nucleases
Zinc-finger nucleases (ZFNs) are engineered endonucleases containing separate domains for DNA-binding and DNA cleavage. Zinc finger (Cys2His2) consists of 30 amino acids forming two β-sheets opposing an α-helix, stabilized by two cysteines and two histidine residues binding a zinc ion, thereby forming a compact globular domain (22). This motif uses residues in the α-helix to bind to approximately three specific base pairs in the major groove of the DNA (23). The ZFN consists of a site-specific zinc-finger DNA-binding domain fused with a nonspecific cleavage domain, generally the FokI nuclease. Since the FokI nuclease functions as a dimer to cleave double-strand DNA, at least two ZFN molecules are required for efficient DNA cleavage at both strands (i.e., 24-36 nucleotides).
Two zinc-finger molecules bind opposite strands of DNA in the tight space, and the FokI nucleases dimerize, cleaving the DNA at the target loci (Figure 2A). The ZFNs have been widely applied in biotechnology and medicine to modify the genomes of many eukaryote cells or organisms, including humans, plants, and Drosophila melanogaster. This strategy has facilitated the progress of targeted gene therapy in humans, and off-target effects appear to be minimal with high-quality ZFN designs. Because ZFNs have minimal target sequence constraints, they can be designed for most genes and regions of the genome (24, 25).
5.2. Transcription Activator-Like Effector Nucleases
Transcription activator-like effectors (TALEs) are plant pathogenic bacteria proteins used by Xanthomonas spp. to modulate gene transcription in host plants to facilitate bacterial colonization. Transcription activator-like effector nucleases (TALENs) are hybrid molecules consisting of central repeat domain and tandem repeats of 34-aminoacid sequences (i.e., termed monomers) required for DNA recognition and binding, fused with the FokI endonuclease catalytic domain (26, 27). The twelfth and thirteenth amino acids of each repeat are highly variable, called repeat variable di-residues, which are specific for the targeted DNA recognition and provide a possibility to design TALE sequences targeting virtually any selected gene (Figure 2A).
Due to their simple protein-DNA code and modular nature, TALENs theoretically can be easily and rapidly constructed to target any sequence and have already been successfully established in many organisms. Because the FokI catalytic domain must dimerize to become active and cleavage DNA, two molecules of TALE nucleases are required to recognize adjacent sequences in both strands of DNA (28, 29). The TALENs exhibit low and minimal off-target effects and cytotoxicity, compared to ZFNs, making them an efficient genome-editing tool (30, 31).
5.3. Clustered Regularly Interspaced Short Palindromic Repeats
Clustered regularly interspaced short palindromic repeats (CRISPR) is the adaptive immune system of bacteria protecting them against foreign genetic fragments through two classes of ribonucleic acid (RNA)-guided DNA endonuclease effects (Figure 2A). Class 1 effectors use multi-protein complexes. On the other hand, class 2 effectors are dependent on single-component effector proteins (32). Almost all archaea and many bacteria achieve adaptive immunity through a diverse set of CRISPR-CAS systems, each containing a combination of CAS proteins and CRISPR RNAs. The CRISPR and its associated proteins are intended for targeted genome modification in human cells and other organisms and are extensively used in research, medicine, and biotechnology (33).
6. Targeting MHC Locus to Provide Histocompatibility
The MHC mismatches between host and donor are important factors in both graft-versus-host and host-versus-graft reactions. The host immune T cell could recognize MHC I and II on the surface of allogeneic T cells. In some cases, pre-existing antibodies could bind to these molecules and mediate the immediate immune rejection of allogeneic cells (34). Conditional therapy resulting in intensive lymphodepletion may be sufficient to allow CAR T-cells to expand and survive before host immune recovery. However, several strategies are deployed to induce the resistance of CAR-T cells to lymphodepletion agents (12).
Alemtuzumab is a humanized immunoglobulin monoclonal antibody binding to CD52 that is expressed in more than 95% of all human blood lymphocytes and most of B- and T-cell lymphomas (35). Monoclonal therapy with alemtuzumab can result in lymphodepletion lasting for weeks; however, engineered T cells with the lack of CD52 expression acquire a survival advantage in the presence of alemtuzumab reducing host-versus-graft reactions (12).
Another approach is the disruption of MHC expression on allogeneic T cells (Figure 2B). Mutation in tapasin, TAP1, or TAP2 (i.e., transporter associated with antigen presentation), results in reduced MHC I expression (36); however, disruption in B2M could prevent the expression of MHC II, which increased expression in activated T cells (37). The other molecules are regulatory factor X complex (i.e., RFX5, RFXB/ANK, and RFXAP) that could be important in MHC II expression. The importance of these strategies is “missing self” responses by host natural killer cells that recognize any cells with the reduced expression of non-polymorphic MHC class I molecules, such as HLA-E providing inhibitory signals (38, 39).
7. Targeting TCR to Overcome Alloreactivity
To prevent the alloreactivity of the T-cell product, endogenous TCR can be selectively deleted by genome-editing techniques to gain off-the-shelf engineered T Cell therapy. Various studies have shown that insertion-deletion at the TCR constant region coding locus can cause endogenous TCR knockout (Figure 2B). The only degradation of the TCRɑ constant chain can cause complete loss of endogenous TCRɑβ function (40). Several studies have shown that knocking out the T-cell receptor alpha constant (TRAC) locus, which is the encoding locus of the constant region of the TCR ɑ chain, can lead to a lack of TCR expression at the surface of engineered cells.
Torikai et al. developed specific universal T cells for tumor-related antigens from a donor to meet the need for patient-specific T cells to transmit them to multiple receptors. This was performed by ZFN gene-editing technique in CD19-specific CAR T cells to eliminate the expression of endogenous TCRs for the prevention of GVHD. The fixed region of alpha or beta TCR chains was targeted by the ZFN technique to eliminate the expression of endogenous TCRs on the surface of engineered cells (41). In 2017, Ren et al., using the CRISPR/Cas9 technique, produced CAR T cells inefficient in expressing endogenous TCR and HLA class 1. In the aforementioned study, the constant region of TCR chains was also targeted by guide RNA electroporation (11).
Eyquem et al. demonstrated that inserting the CAR in the TRAC locus using the CRISPR/Cas9 genome-editing system increases tumor killing by the CAR T cells. The insertion of CD19-specific CAR in the TRAC locus leads to uniform CAR expression in human peripheral blood T cells and increases the potency of T cells. Eyquem et al. also showed that inserting the CAR at the TRAC locus would effectively enter and re-express the CAR after only one exposure or repeated exposure to antigen, as well as delayed dissociation and exhausting of effector T cells. The aforementioned findings revealed the immunological principle of CAR. To knock out the TRAC locus and insert the CD19-specific CAR in this locus, this group designed a guide RNA that targets the 5' end of the first TRAC exon (42).
MacLeod et al. indicated that the integration of CD19-CAR into the TCR’s alpha chain locus facilitates the allogeneic production of engineered CAR T cells. In these cells produced by the TALEN genome-editing system, endogenous TCR was not expressed. These cells showed vital in vitro effector function and clearance of CD19+ tumors in the mice model (43).
In 2017, Qasim et al. reported the first human administration of genome-edited T cells for two infants with B-cell acute lymphoblastic leukemia. In this clinical trial, universal CAR19 cells were manufactured from HLA-incompatible donor cells. Universal CAR19 cells, using the TALEN technique simultaneously, intervene in the expression of endogenous CD52 and TCR, one to prevent the destructive effects of alemtuzumab and the other to reduce GVHD, in which the successful induction of molecular remission was observed before allogeneic stem cell transplantation. The aforementioned clinical trial also targeted the constant region of the TCR alpha chain to access engineered cells by removing endogenous TCR. Therefore, it was demonstrated that it was possible to produce a universal CAR T-cell bank (44). Since then, further clinical trials have been conducted on genome-edited T cell therapies. These ongoing trials are expected to result in a new generation of off-the-shelf T-cell therapies.
8. Conclusions
Genetically engineered T cells, including CAR and TCR, pave their way into clinical practice. However, the current model of autologous patient-specific manufacturing limits the application of these therapies. Genome-editing technologies are promising means to tackle these limitations and provide an off-the-shelf model for these cell-based immunotherapies. Current preclinical and clinical reports have demonstrated the potential of these technologies for the generation of genetically engineered allogeneic T cells. This trend suggests that the next generation of CAR and TCR-engineered T cell therapies for cancer will rely on genome-edited off-the-shelf T cells.
References
-
1.
Kershaw MH, Westwood JA, Darcy PK. Gene-engineered T cells for cancer therapy. Nat Rev Cancer. 2013;13(8):525-41. doi: 10.1038/nrc3565. [PubMed: 23880905].
-
2.
Knochelmann HM, Smith AS, Dwyer CJ, Wyatt MM, Mehrotra S, Paulos CM. CAR T cells in solid tumors: Blueprints for building effective therapies. Front Immunol. 2018;9:1740. doi: 10.3389/fimmu.2018.01740. [PubMed: 30140266]. [PubMed Central: PMC6094980].
-
3.
Smith TWJ, Nishimura MI. Targeting cancer with genetically engineered TCR T Cells. Recent Results Cancer Res. 2020;214:129-51. doi: 10.1007/978-3-030-23765-3_4. [PubMed: 31473851].
-
4.
Ellis GI, Sheppard NC, Riley JL. Genetic engineering of T cells for immunotherapy. Nat Rev Genet. 2021;22(7):427-47. doi: 10.1038/s41576-021-00329-9. [PubMed: 33603158]. [PubMed Central: PMC8217325].
-
5.
Ikeda H. T-cell adoptive immunotherapy using tumor-infiltrating T cells and genetically engineered TCR-T cells. Int Immunol. 2016;28(7):349-53. doi: 10.1093/intimm/dxw022. [PubMed: 27127191].
-
6.
Bastien JP, Minguy A, Dave V, Roy DC. Cellular therapy approaches harnessing the power of the immune system for personalized cancer treatment. Semin Immunol. 2019;42:101306. doi: 10.1016/j.smim.2019.101306. [PubMed: 31604534].
-
7.
Themeli M, Riviere I, Sadelain M. New cell sources for T cell engineering and adoptive immunotherapy. Cell Stem Cell. 2015;16(4):357-66. doi: 10.1016/j.stem.2015.03.011. [PubMed: 25842976]. [PubMed Central: PMC5611846].
-
8.
Ping Y, Liu C, Zhang Y. T-cell receptor-engineered T cells for cancer treatment: current status and future directions. Protein Cell. 2018;9(3):254-66. doi: 10.1007/s13238-016-0367-1. [PubMed: 28108950]. [PubMed Central: PMC5829268].
-
9.
Campillo-Davo D, Flumens D, Lion E. The quest for the best: How TCR affinity, avidity, and functional avidity affect TCR-engineered T-cell antitumor responses. Cells. 2020;9(7). doi: 10.3390/cells9071720. [PubMed: 32708366]. [PubMed Central: PMC7408146].
-
10.
Chmielewski M, Hombach AA, Abken H. Antigen-specific T-cell activation independently of the MHC: Chimeric antigen receptor-redirected T cells. Front Immunol. 2013;4:371. doi: 10.3389/fimmu.2013.00371. [PubMed: 24273543]. [PubMed Central: PMC3822734].
-
11.
Ren J, Liu X, Fang C, Jiang S, June CH, Zhao Y. Multiplex genome editing to generate universal CAR T cells resistant to PD1 inhibition. Clin Cancer Res. 2017;23(9):2255-66. doi: 10.1158/1078-0432.CCR-16-1300. [PubMed: 27815355]. [PubMed Central: PMC5413401].
-
12.
Poirot L, Philip B, Schiffer-Mannioui C, Le Clerre D, Chion-Sotinel I, Derniame S, et al. Multiplex genome-edited T-cell manufacturing platform for "Off-the-shelf" adoptive T-cell immunotherapies. Cancer Res. 2015;75(18):3853-64. doi: 10.1158/0008-5472.CAN-14-3321. [PubMed: 26183927].
-
13.
Sharpe M, Mount N. Genetically modified T cells in cancer therapy: opportunities and challenges. Dis Model Mech. 2015;8(4):337-50. doi: 10.1242/dmm.018036. [PubMed: 26035842]. [PubMed Central: PMC4381333].
-
14.
Rafiq S, Hackett CS, Brentjens RJ. Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat Rev Clin Oncol. 2020;17(3):147-67. doi: 10.1038/s41571-019-0297-y. [PubMed: 31848460]. [PubMed Central: PMC7223338].
-
15.
Townsend MH, Bennion K, Robison RA, O'Neill KL. Paving the way towards universal treatment with allogenic T cells. Immunol Res. 2020;68(1):63-70. doi: 10.1007/s12026-020-09119-7. [PubMed: 32096010].
-
16.
Depil S, Duchateau P, Grupp SA, Mufti G, Poirot L. 'Off-the-shelf' allogeneic CAR T cells: development and challenges. Nat Rev Drug Discov. 2020;19(3):185-99. doi: 10.1038/s41573-019-0051-2. [PubMed: 31900462].
-
17.
Robinson J, Halliwell JA, Hayhurst JD, Flicek P, Parham P, Marsh SG. The IPD and IMGT/HLA database: Allele variant databases. Nucleic Acids Res. 2015;43(Database issue):D423-31. doi: 10.1093/nar/gku1161. [PubMed: 25414341]. [PubMed Central: PMC4383959].
-
18.
Archbold JK, Ely LK, Kjer-Nielsen L, Burrows SR, Rossjohn J, McCluskey J, et al. T cell allorecognition and MHC restriction--A case of Jekyll and Hyde? Mol Immunol. 2008;45(3):583-98. doi: 10.1016/j.molimm.2006.05.018. [PubMed: 17869342].
-
19.
Gokmen MR, Lombardi G, Lechler RI. The importance of the indirect pathway of allorecognition in clinical transplantation. Curr Opin Immunol. 2008;20(5):568-74. doi: 10.1016/j.coi.2008.06.009. [PubMed: 18655831].
-
20.
Kim DW, Cho JY. Recent advances in allogeneic CAR-T cells. Biomolecules. 2020;10(2). doi: 10.3390/biom10020263. [PubMed: 32050611]. [PubMed Central: PMC7072190].
-
21.
Delhove J, Qasim W. Genome-edited T cell therapies. Curr Stem Cell Rep. 2017;3(2):124-36. doi: 10.1007/s40778-017-0077-5. [PubMed: 28596938]. [PubMed Central: PMC5445182].
-
22.
Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem. 2001;70:313-40. doi: 10.1146/annurev.biochem.70.1.313. [PubMed: 11395410].
-
23.
Pavletich NP, Pabo CO. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252(5007):809-17. doi: 10.1126/science.2028256. [PubMed: 2028256].
-
24.
Petersen B, Niemann H. Advances in genetic modification of farm animals using zinc-finger nucleases (ZFN). Chromosome Res. 2015;23(1):7-15. doi: 10.1007/s10577-014-9451-7. [PubMed: 25596823].
-
25.
Davies JP, Kumar S, Sastry-Dent L. Use of zinc-finger nucleases for crop improvement. Prog Mol Biol Transl Sci. 2017;149:47-63. doi: 10.1016/bs.pmbts.2017.03.006. [PubMed: 28712500].
-
26.
Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7(1):171-92. doi: 10.1038/nprot.2011.431. [PubMed: 22222791]. [PubMed Central: PMC3684555].
-
27.
Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419-36. doi: 10.1146/annurev-phyto-080508-081936. [PubMed: 19400638].
-
28.
Lainscek D, Lebar T, Jerala R. Transcription activator-like effector-mediated regulation of gene expression based on the inducible packaging and delivery via designed extracellular vesicles. Biochem Biophys Res Commun. 2017;484(1):15-20. doi: 10.1016/j.bbrc.2017.01.090. [PubMed: 28111345].
-
29.
Sun N, Zhao H. Transcription activator-like effector nucleases (TALENs): A highly efficient and versatile tool for genome editing. Biotechnol Bioeng. 2013;110(7):1811-21. doi: 10.1002/bit.24890. [PubMed: 23508559].
-
30.
Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39(21):9283-93. doi: 10.1093/nar/gkr597. [PubMed: 21813459]. [PubMed Central: PMC3241638].
-
31.
Menoret S, Tesson L, Remy S, Usal C, Thepenier V, Thinard R, et al. Gene targeting in rats using transcription activator-like effector nucleases. Methods. 2014;69(1):102-7. doi: 10.1016/j.ymeth.2014.02.027. [PubMed: 24583114].
-
32.
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759-71. doi: 10.1016/j.cell.2015.09.038. [PubMed: 26422227]. [PubMed Central: PMC4638220].
-
33.
Rosenblum D, Gutkin A, Dammes N, Peer D. Progress and challenges towards CRISPR/Cas clinical translation. Adv Drug Deliv Rev. 2020;154-155:176-86. doi: 10.1016/j.addr.2020.07.004. [PubMed: 32659256].
-
34.
Morin-Zorman S, Loiseau P, Taupin JL, Caillat-Zucman S. Donor-specific anti-HLA antibodies in allogeneic hematopoietic stem cell transplantation. Front Immunol. 2016;7:307. doi: 10.3389/fimmu.2016.00307. [PubMed: 27570526]. [PubMed Central: PMC4981597].
-
35.
Salisbury JR, Rapson NT, Codd JD, Rogers MV, Nethersell AB. Immunohistochemical analysis of CDw52 antigen expression in non-Hodgkin's lymphomas. J Clin Pathol. 1994;47(4):313-7. doi: 10.1136/jcp.47.4.313. [PubMed: 8027367]. [PubMed Central: PMC501933].
-
36.
Yabe T, Kawamura S, Sato M, Kashiwase K, Tanaka H, Ishikawa Y, et al. A subject with a novel type I bare lymphocyte syndrome has tapasin deficiency due to deletion of 4 exons by Alu-mediated recombination. Blood. 2002;100(4):1496-8. doi: 10.1182/blood-2001-12-0252. [PubMed: 12149238].
-
37.
Riolobos L, Hirata RK, Turtle CJ, Wang PR, Gornalusse GG, Zavajlevski M, et al. HLA engineering of human pluripotent stem cells. Mol Ther. 2013;21(6):1232-41. doi: 10.1038/mt.2013.59. [PubMed: 23629003]. [PubMed Central: PMC3677304].
-
38.
Pallmer K, Oxenius A. Recognition and regulation of T cells by NK cells. Front Immunol. 2016;7:251. doi: 10.3389/fimmu.2016.00251. [PubMed: 27446081]. [PubMed Central: PMC4919350].
-
39.
Gornalusse GG, Hirata RK, Funk SE, Riolobos L, Lopes VS, Manske G, et al. HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat Biotechnol. 2017;35(8):765-72. doi: 10.1038/nbt.3860. [PubMed: 28504668]. [PubMed Central: PMC5548598].
-
40.
Alcover A, Alarcon B, Di Bartolo V. Cell biology of T cell receptor expression and regulation. Annu Rev Immunol. 2018;36:103-25. doi: 10.1146/annurev-immunol-042617-053429. [PubMed: 29261409].
-
41.
Torikai H, Reik A, Liu PQ, Zhou Y, Zhang L, Maiti S, et al. A foundation for universal T-cell based immunotherapy: T cells engineered to express a CD19-specific chimeric-antigen-receptor and eliminate expression of endogenous TCR. Blood. 2012;119(24):5697-705. doi: 10.1182/blood-2012-01-405365. [PubMed: 22535661]. [PubMed Central: PMC3382929].
-
42.
Eyquem J, Mansilla-Soto J, Giavridis T, van der Stegen SJ, Hamieh M, Cunanan KM, et al. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543(7643):113-7. doi: 10.1038/nature21405. [PubMed: 28225754]. [PubMed Central: PMC5558614].
-
43.
MacLeod DT, Antony J, Martin AJ, Moser RJ, Hekele A, Wetzel KJ, et al. Integration of a CD19 CAR into the TCR alpha chain locus streamlines production of allogeneic gene-edited CAR T cells. Mol Ther. 2017;25(4):949-61. doi: 10.1016/j.ymthe.2017.02.005. [PubMed: 28237835]. [PubMed Central: PMC5383629].
-
44.
Qasim W, Zhan H, Samarasinghe S, Adams S, Amrolia P, Stafford S, et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci Transl Med. 2017;9(374). doi: 10.1126/scitranslmed.aaj2013. [PubMed: 28123068].