Abstract
Background:
Managing individual-based anti-coagulant therapy with warfarin is an important goal for bleeding prevention in personalized medicine.Objectives:
The purpose of the present study was to evaluate the allelic frequency of VKORC1 (C1173T, G1639A) and CYP2C9 (CYP2C9*2 and CYP2C9*3) polymorphic genes with SimpleProbe® real-time PCR as a fast, accurate and easy-to-handle method.Patients and Methods:
One hundred unrelated patients under warfarin therapy were recruited as the study population. Real-time PCR was performed with SimpleProbe® for single-nucleotide polymorphisms (SNP) detection of CYP2C9 and VKORC1 genes.Results:
The allelic frequencies for VKORC1 C1173T (CC, CT, TT) were 25%, 51% and 24%, respectively and for VKORC1 G1639A (-GG, GA, AA) were 27%, 51% and 22%, respectively. Despite a significant association between allelic polymorphisms in VKORC1 and warfarin dose (P < 0.001), it was not statistically significant for CYP2C9*2 and CYP2C9*3, most probably due to the low rate of CYP2C9*2 and CYP2C9*3 SNPs observed.Conclusions:
SimpleProbe® real-time PCR is a fast and accurate technique qualified to detect VKORC1 and CYP2C9 SNPs. These results encourage taking further steps towards using VKORC1 and CYP2C9 allelic screening to prevent clinical complications due to resistance or sensitivity to warfarin as well as anti-coagulant dose adjustment.Keywords
VKORC1 CYP2C9 SNP Real-Time Polymerase Chain Reaction Warfarin
1. Background
Warfarin (Coumadin) was introduced into clinical practice in the 1950s, and has been the most widely prescribed oral anti-coagulant for prevention and treatment of thrombotic diseases despite its significant side effects (1-3). The small therapeutic window of warfarin necessitates individual dosing for bleeding risks (4, 5).
Several clinical variables are considered for initiating the warfarin therapy. Although the inter-individual variability depends on environmental factors, the effect of genetic variations has also been demonstrated, and increasing evidence supports the association of genotype and bleeding tendency following warfarin therapy (6).
Vitamin K epoxide reductase complex subunit1 (VKORC1), a membrane protein located in hepatocytes, has an important role in the anabolism of vitamin K which is essential for blood coagulation. Coumarin derivatives like warfarin act as vitamin K antagonists (VKA) by inhibiting the VKORC1 activity and thus, regeneration of reduced vitamin K. Recently reported polymorphisms in the VKORC1 gene, justifies up to 30% of the variability in the pharmacological responses to VKAs. A VKORC1-1639 AA genotype carrier requires a significantly lower daily dose of warfarin than GA or GG genotype carriers (7-11).
The VKORC1, 1639G > A and 1173C > T polymorphisms have strong linkage disequilibrium and show a significant effect on the response to a given dose of warfarin.
Termination of the warfarin pharmacological effects depends on hepatic metabolism, catalyzed largely by the cytochrome P450 (CYP) system. This enzyme family consists of more than 30 isoenzymes mediating drug metabolism like S-warfarin. CYP2C9 is the most abundant enzyme among human CYP2C isoforms primarily localized in liver accounting for 20% of the total hepatic CYP450 content. The CYP2C9 genetic polymorphisms are associated with variations in the levels of enzymatic activity. Among all allelic variants, CYP2C9*2 and CYP2C9*3 show clinical significance and are associated with decreased ability to metabolize the substrates in comparison with the common wild-type CYP2C9*1. Among the Caucasians, the most common mutant allele is CYP2C9*2 (8 - 12%) causing 30–50% lower turnover of S-warfarin, while the frequency of CYP2C9*3 varies from 5 to 9% (7-12) causing a much lower intrinsic clearance (approximately 90%), which means patients carrying these polymorphisms metabolize warfarin at a slower rate and may be at a greater risk of bleeding during initiation of traditional warfarin therapy.
Despite numerous worldwide studies, only a few have investigated the distribution frequency of major allelic variations and their effects on drug sensitivity and metabolism among Iranian population (7, 8, 12-14) using time-consuming and technically cumbersome approaches (8, 14, 15). Therefore, more robust SNP detection methods with higher accuracy are essential for screening purposes. Real-time PCR with SimpleProbe® format for SNP genotyping and mutation detection is a fast, reliable, sensitive and easy-to-handle technique applied in this investigation.
Single-labeled (SimpleProbe®) is a sequence specific hybridization probe, which fluoresces when bound to a single DNA strand. As a result, changes in fluorescence are solely based on the hybridization status of the probe. By measuring the fluorescence, the instrument can detect melting of the probe-target hybrids by increasing the temperature. A mismatch (like SNPs) weakens the stability of the SimpleProbe® and prompts probe melting at a lower temperature compared to perfectly matched sequence, leading to detection of the mutation.
2. Objectives
This study was designed to evaluate the allelic frequency of the VKORC1 (C1173T, G1639A) and CYP2C9 (CYP2C9*2 and CYP2C9*3) polymorphic genes with SimpleProbe® real-time PCR.
3. Patients and Methods
3.1. Patients
One hundred unrelated patients with heart failure history merely under warfarin therapy were enrolled in this study during 2010 - 2012. The subjects were referred to the Laboratory of Day General Hospital (Tehran, Iran) for routine laboratory tests and then underwent the two common laboratory tests used to evaluate coagulation disorders including Prothrombin Time (PT) and Partial Thromboplastin Time (PTT). The PT and international normalized ratio (INR) are used to monitor the effectiveness of warfarin. Each manufacturer assigns an international sensitivity index (ISI) value (usually between 1.0 and 2.0) for any manufactured tissue factor which indicates how a particular batch compares to an international reference tissue factor. The INR is the ratio of a patient's prothrombin time to a normal (control) sample, raised to the power of the ISI value for the analytical system used. Hence, INR is calculated as (PT test/PT control) ISI.
The PT and PTT were determined using standard Kits (HemosIL TM, Instrumentation Laboratory Company, USA) according to the manufacturer’s instruction. All procedures were approved by an Ethics Committee, and participants' consent was obtained.
3.2. DNA Extraction and SimpleProbe® Real-Time PCR
Blood samples were collected in tubes containing EDTA. DNA extraction was performed using QIAamp Blood DNA Mini Kit (Qiagen, USA) according to the manufacturer’s manual, with final elution in 50 µL of nuclease-free water. The prepared DNA was assayed immediately or stored at -20ºC until being used.
SimpleProbe® real-time PCR assay for CYP2C9*2 and CYP2C9*3 was performed using light Mix® Kit human CYP2C9*2 and CYP2C9*3 (TIB MOLBIOL GmbH, Berlin, Germany) in 10 µL PCR mixture volume composed of 2 µL Light Cycler® Fast Start DNA Master HybProbe® (Roche Diagnostics, USA), 2 µL Primer and Probe (TIB MOLBIOL GmbH, Berlin, Germany), and 2.5 µL extracted DNA. Amplification was performed in the following conditions: activation step at 95°C for 10 minutes and 45 cycles of three thermal amplification steps: 95°C for 5 seconds, 60°C for 10 seconds, and 72°C for 15 seconds. The genotypes were identified by running a melting curve with specific Tm of 58.5ºC for the wild type and 50.5°C for the mutant human CYP2C9*2 in channel 530 (F1) and 48.5°C for the wild type and 58.3°C for the mutant human CYP2C9*3 in channel 640 (F2). Amplification, data acquisition and melting curve analysis were performed on Light cycler® 1.5 (Roche, USA).
Real time PCR assay for VKORC1 was performed using light Mix® for detection of VKORC1 C1173T and VKORC1 G1639A (TIB MOLBIOL GmbH, Berlin, Germany) according to the manufacturer’s protocol. The wild type VKORC1 C1173 and G1639 exhibited a Tm of 51.8°C in channel F1 and a Tm of 52.6°C in channel F2, respectively. The allele variants VKORC1 1173T and 1639A exhibited a Tm of 58.1°C in channel F1 and a Tm of 61.3°C in channel F2. The assay sensitivity was 1 ng, and measuring range of the assay was 1 - 100 ng of genomic DNA.
3.3. Statistics
SPSS version 16.0 was used for data analysis. The Kolmogorov-Smirnov test was used to determine the normal distribution of the values and to indicate whether parametric or nonparametric statistical analysis was required for analyzing the test results. One-way analysis of variance (ANOVA) was used to assess group differences for continuous variables and χ2 test for categorical variables. Descriptive test was used to obtain the frequencies of all parameters in the study population. A P value < 0.05 was considered statistically significant.
4. Results
This study was performed on one hundred ward patients with heart condition, referred to the Laboratory of Day General Hospital for PT, PPT and INR assessments. These patients were screened to be under warfarin therapy as the single anticoagulant therapy. Mutation detection for VKORC1 and CYP2CP was performed on this group regarding the issue that the pharmacodynamic mechanism of warfarin resistance relies on the CYP2C9 or VKORC1 polymorphism.
Previous reports showed that the prevalence of VKORC1 and CYP2C9 mutations differ between the ethnic groups. African-Americans have a higher proportion of the wild-type genotype for both CYP2C9 and VKORC1 compared to Caucasians and Asians. CPY2C9*2 variant is found more commonly in Caucasians, while CYP2C9*3 is common in Asians (16).
SimpleProbe® real-time PCR assay was used to detect CYP2C9*2, CYP2C9*3, VKORC1 G1639A and VKORC1 C1173T alleles in the study group as a representative of the Iranian population. Schematic results of genotyping by real-time PCR are shown in Figure 1. SimpleProbe® real-time PCR had sufficient accuracy to detect wild type, heterozygote and mutant genotypes in the study population based on their Tm.
Schematic Representation of Genotyping Pattern by SimpleProbe® Real-Time PCR
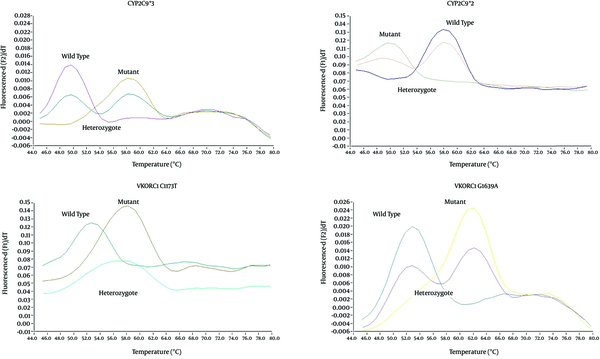
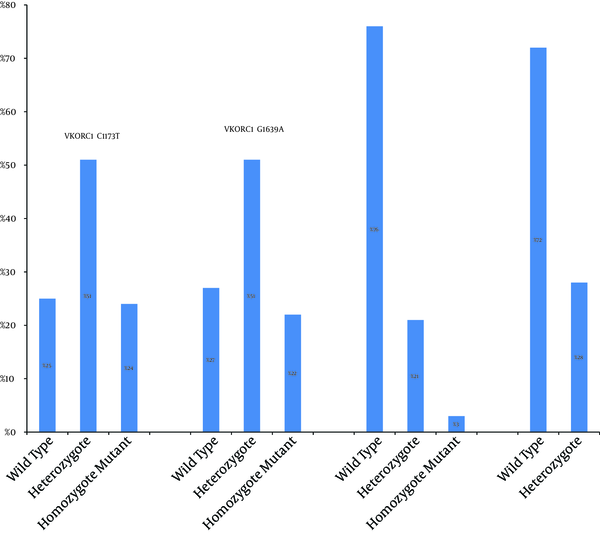
As demonstrated in genotyping patterns in Figure 2, VKORC1 1639-GA was the most abundant genotype of VKORC1 1639 allele. The two homozygote -GG and -AA genotypes had frequencies of 27% and 22%, respectively. In SimpleProbe® real-time PCR of VKORC1 C1173T, heterozygote genotype was more common and accounted for 51% of the samples. The two homozygote -CC and -TT genotypes had frequencies of 25% and 24%, respectively. Thus, in both VKORC1 (C1173T and G1639A) genes, heterozygote genotype was more common in a total of 100 patients. In CYP2C9 variant, 21 and 28 subjects carried CYP2C9*1/*2 and CYP2C9*1/*3, respectively. Three individuals (3%) had the genotype CYP2C9*2/*2, while CYP2C9*3/*3 was not detected in any of the samples (Figure 2). Therefore, wild type genotype was dominant in both CYP2C9 (*2 or *3) genes.
Allelic Frequencies for VKORC1 C1173T, VKORC1 G1639A, CYP2C9*2, and CYP2C9*3 in 100 Patients Enrolled in This Study
In the studied population, the average age was 57.15 ± 17.34 SD ranging from 11 to 84 years old. The mean ± standard error (SE) of warfarin maintenance dose was 28.25 ± 0.33 mg /week. INR mean ± SE was 2.30 ± 0.56 ranging from 1.0 to 3.7. Test of normality with Kolmogorov-Smirnov demonstrated that PT, PTT, INR and warfarin dose data had normal distribution. One-way ANOVA test indicated a significant association between drug dose and both VKORC1 C1173T and VKORC1 G1639A. No significant association was found between CYP2C9*2, CYP2C9*3 genotypes and INR or drug dose.
5. Discussion
Warfarin and related anticoagulants are widely prescribed drugs to prevent coagulation in cardiovascular problems. The drug's small therapeutic window requires improving the safety of oral anticoagulation administration (6, 16-19). Most of the inter-individual and interethnic differences in the effects of drugs are attributed to genetic differences in their metabolism. Although the dosage of warfarin is currently adjusted according to the INR, a significant inter-individual variability exists in the dose-response relationship, leading to bleeding and insufficient anticoagulation as adverse responses (5, 6, 16).
The first study on the role of VKORC1 genetic polymorphisms in setting of the required warfarin dose was published in 2005 by D’Andrea et al. (9). The required dose of an antagonist to achieve the target level of anticoagulation in VKA therapy is variable in individuals and depends on numerous factors such as age, dietary intake, variations in pharmacokinetics as well as interfering pharmacotherapy (20). Each population has a unique pattern of gene polymorphism, playing a significant role in drug metabolism (6). As demonstrated in Table 1, the results of the present study on Iranian population represented that patients with one or two mutated alleles in VKORC1 C1173T and VKORC1 G1639A required lower warfarin maintenance doses. In this study, the frequency of VKORC1-1639A polymorphism (51%) was similar to what previously reported for the Caucasians, but it was different from the African-American and Chinese populations (13% and 96%, respectively) (7).
Warfarin Maintenance Doses in Patients With Allelic Diversities in VKORC1 C1173T, VKORC1 G1639A, CYP2C9*2 and CYP2C9*3 Genes a
| Gene | SNP | Warfarin Dose ± SEM, mg/wk | P value |
|---|---|---|---|
| VKORC1 1173 | CC | 7.62 ± 0.81 | < 0.001 |
| CT | 5.48 ± 0.39 | ||
| TT | 3.9 ± 0.45 | ||
| VKORC1 1639 | GG | 7.35 ± 0.77 | < 0.001 |
| GA | 5.57 ± 0.40 | ||
| AA | 3.67 ± 0.38 | ||
| CYP2C9*2 | *1/*1 | 5.81 ± 0.37 | 0.68 |
| *1/*2 | 5.18 ± 0.79 | ||
| *2/*2 | 4.8 ± 0.82 | ||
| CYP2C9*3 | *1/*1 | 5.84 ± 0.37 | 0.365 |
| *1/*3 | 5.17 ± 0.69 |
The frequency of VKORC1-1639 AA in Asians, African-Americans and Europeans were 92%, 0% and 23%, respectively. But according to the previous reports in the UK, France and Iran, the related frequencies were 19%, 23% and 27% close to the findings of the present study (22%) (21, 22).
Some genetic polymorphisms of CYP2C9 leading to changes in amino acid sequence of these enzymes can alter both enzymatic activity and substrate specificity. Human CYP2C9*2 is the most clinically important allele of CYP2C9 in Caucasians (11, 15). Herein, the presence of genetic heterozygosis in CYP 2C9 was demonstrated. Wild type genotype (CYP2C91*1) has been reported in about 75% of the Iranians (7) which is in accordance with our results and slightly different from reports from Egypt (23) and India (82%) (24). Three patients with homozygote state for CYP2C9*2 allele were found to have warfarin maintenance dose of 24 mg/week which is lower than the average warfarin prescribed dose (28.25 ± 0.33 mg/week). No case of homozygote state for CYP2C93*3 genotype was verified in the studied population, which is in accordance with previous studies in Iran (15) as well as some other reports from Greece (25) and Italy (26). The frequency of CYP2C91*3 is very low (< 8%) in most populations such as the Italians (26), Swedish (27), Slovenians (28), Egyptians (23) Indians (24), and Russians (29).
Real-time PCR using SimpleProbe®, SYBR-Green I (30), PCR-RFLP and smart amplification process (31) are excellent tools for SNP genotyping and mutation detection. Several companies are developing commercial rapid genotyping assays for CYP2C9 and VKORC1 based on microchips, bead technology or fluoresceinated DNA probes to identify biotinylated amplicons (2). SimpleProbe® is more accurate and facilitates readily identification of wild-type, mutant, and heterozygous samples with only a single short probe (4, 32-34). The Light Cycler® Instrument recruited in this study is capable of monitoring the melting behavior of the SimpleProbe®. To this date, few studies have determined the frequency of CYP2C9*2, CYP2C9*3 or VKORC1 allelic variants among the Iranian population (7, 8, 13, 15, 16).
According to previous findings, polymorphisms of CYP2C9 and VKORC1 are associated with variability in response to warfarin. Identifying these polymorphisms in patients prior to starting the therapy may help clinicians choose the appropriate strategy to reduce adverse drug reactions such as bleeding. The low rate of mutated CYP2C9*2 and CYP2C9*3 alleles reported in this study, makes it difficult to conclude an accurate association between allelic variation and warfarin dosage, and should be further investigated using larger samples. It may be concluded that prospective application of SimpleProbe® real-time PCR for VKORC1 and CYP2C9 polymorphism status detection as an approach for settling warfarin dosage can be accurate, technically feasible and acceptable for patients and service providers.
Individualized medicine is becoming a reality in future medicine. Clinicians should not ignore the large amount of new information pertaining to pharmacogenetic testing and should apply it in the already complex process of individual therapeutic decision-making particularly when the patients’ safety is a matter of concern. Use of genetic polymorphisms for CYP2C9 and VKORC1, environmental and clinical factors to estimate the warfarin dosage could potentially minimize the risk of overdose during warfarin therapy. Testing for VKORC1 polymorphism complemented with testing for CYP2C9 polymorphisms, may become one of the necessary standards for all patients prior to starting warfarin therapy.
Acknowledgements
References
-
1.
Cavallari LH, Limdi NA. Warfarin pharmacogenomics. Curr Opin Mol Ther. 2009;11(3):243-51. [PubMed ID: 19479657].
-
2.
Gage BF. Pharmacogenetics-based coumarin therapy. Hematology Am Soc Hematol Educ Program. 2006:467-73. [PubMed ID: 17124101]. https://doi.org/10.1182/asheducation-2006.1.467.
-
3.
Hall AM, Wilkins MR. Warfarin: a case history in pharmacogenetics. Heart. 2005;91(5):563-4. [PubMed ID: 15831631]. https://doi.org/10.1136/hrt.2004.051771.
-
4.
Lefferts JA, Schwab MC, Dandamudi UB, Lee HK, Lewis LD, Tsongalis GJ. Warfarin genotyping using three different platforms. Am J Transl Res. 2010;2(4):441-6. [PubMed ID: 20733952].
-
5.
Moyer TP, O'Kane DJ, Baudhuin LM, Wiley CL, Fortini A, Fisher PK, et al. Warfarin sensitivity genotyping: a review of the literature and summary of patient experience. Mayo Clin Proc. 2009;84(12):1079-94. [PubMed ID: 19955245]. https://doi.org/10.4065/mcp.2009.0278.
-
6.
Yin T, Miyata T. Warfarin dose and the pharmacogenomics of CYP2C9 and VKORC1 - rationale and perspectives. Thromb Res. 2007;120(1):1. [PubMed ID: 17161452]. https://doi.org/10.1016/j.thromres.2006.10.021.
-
7.
Azarpira N, Namazi S, Hendijani F, Banan M, Darai M. Investigation of allele and genotype frequencies of CYP2C9, CYP2C19 and VKORC1 in Iran. Pharmacol Rep. 2010;62(4):740-6. [PubMed ID: 20885015].
-
8.
Baniasadi S, Beizaee S, Kazemi B, Behzadnia N, Shafaghi B, Bandehpour M, et al. Novel VKORC1 mutations associated with warfarin sensitivity. Cardiovasc Ther. 2011;29(4):e1-5. [PubMed ID: 20579077]. https://doi.org/10.1111/j.1755-5922.2009.00107.x.
-
9.
D'Andrea G, D'Ambrosio RL, Di Perna P, Chetta M, Santacroce R, Brancaccio V, et al. A polymorphism in the VKORC1 gene is associated with an interindividual variability in the dose-anticoagulant effect of warfarin. Blood. 2005;105(2):645-9. [PubMed ID: 15358623]. https://doi.org/10.1182/blood-2004-06-2111.
-
10.
Greer FR. Vitamin K the basics--what's new? Early Hum Dev. 2010;86 Suppl 1:43-7. [PubMed ID: 20116943]. https://doi.org/10.1016/j.earlhumdev.2010.01.015.
-
11.
Pasmant E, de Beauvoir C, Plessier A, Labreuche J, Bezeaud A. VKORC1 and CYP2C9 genetic polymorphisms in hepatic or portal vein thrombosis. Thromb Res. 2010;126(2):e134-6. [PubMed ID: 20434758]. https://doi.org/10.1016/j.thromres.2010.03.015.
-
12.
Zhou SF, Zhou ZW, Huang M. Polymorphisms of human cytochrome P450 2C9 and the functional relevance. Toxicology. 2010;278(2):165-88. [PubMed ID: 19715737]. https://doi.org/10.1016/j.tox.2009.08.013.
-
13.
Ghadam P, Sharifian R, Farsangi ZJ, Kianmehr Z, Lak M. CYP2C9 gene analysis of some Iranian hypersensitive patients to warfarin. Pak J Biol Sci. 2009;12(16):1160-3. [PubMed ID: 19899329].
-
14.
Namazi S, Azarpira N, Hendijani F, Khorshid MB, Vessal G, Mehdipour AR. The impact of genetic polymorphisms and patient characteristics on warfarin dose requirements: a cross-sectional study in Iran. Clin Ther. 2010;32(6):1050-60. [PubMed ID: 20637959]. https://doi.org/10.1016/j.clinthera.2010.06.010.
-
15.
Zand N, Tajik N, Moghaddam AS, Milanian I. Genetic polymorphisms of cytochrome P450 enzymes 2C9 and 2C19 in a healthy Iranian population. Clin Exp Pharmacol Physiol. 2007;34(1-2):102-5. [PubMed ID: 17201743]. https://doi.org/10.1111/j.1440-1681.2007.04538.x.
-
16.
Sinxadi P, Blockman M. Warfarin resistance. Cardiovasc J Afr. 2008;19(4):215-7. [PubMed ID: 18776969].
-
17.
Loeliger EA, Poller L, Samama M, Thomson JM, Van den Besselaar AM, Vermylen J, et al. Questions and answers on prothrombin time standardisation in oral anticoagulant control. Thromb Haemost. 1985;54(2):515-7. [PubMed ID: 4082086].
-
18.
Rettie AE, Farin FM, Beri NG, Srinouanprachanh SL, Rieder MJ, Thijssen HH. A case study of acenocoumarol sensitivity and genotype-phenotype discordancy explained by combinations of polymorphisms in VKORC1 and CYP2C9. Br J Clin Pharmacol. 2006;62(5):617-20. [PubMed ID: 16869821]. https://doi.org/10.1111/j.1365-2125.2006.02688.x.
-
19.
Kimura R, Miyashita K, Kokubo Y, Akaiwa Y, Otsubo R, Nagatsuka K, et al. Genotypes of vitamin K epoxide reductase, gamma-glutamyl carboxylase, and cytochrome P450 2C9 as determinants of daily warfarin dose in Japanese patients. Thromb Res. 2007;120(2):181-6. [PubMed ID: 17049586]. https://doi.org/10.1016/j.thromres.2006.09.007.
-
20.
Yoshizawa M, Hayashi H, Tashiro Y, Sakawa S, Moriwaki H, Akimoto T, et al. Effect of VKORC1-1639 G>A polymorphism, body weight, age, and serum albumin alterations on warfarin response in Japanese patients. Thromb Res. 2009;124(2):161-6. [PubMed ID: 19135231]. https://doi.org/10.1016/j.thromres.2008.11.011.
-
21.
Steimer W, Zopf K, von Amelunxen S, Pfeiffer H, Bachofer J, Popp J, et al. Allele-specific change of concentration and functional gene dose for the prediction of steady-state serum concentrations of amitriptyline and nortriptyline in CYP2C19 and CYP2D6 extensive and intermediate metabolizers. Clin Chem. 2004;50(9):1623-33. [PubMed ID: 15205367]. https://doi.org/10.1373/clinchem.2003.030825.
-
22.
Mushiroda T, Ohnishi Y, Saito S, Takahashi A, Kikuchi Y, Saito S, et al. Association of VKORC1 and CYP2C9 polymorphisms with warfarin dose requirements in Japanese patients. J Hum Genet. 2006;51(3):249-53. [PubMed ID: 16432637]. https://doi.org/10.1007/s10038-005-0354-5.
-
23.
Hamdy SI, Hiratsuka M, Narahara K, El-Enany M, Moursi N, Ahmed MS, et al. Allele and genotype frequencies of polymorphic cytochromes P450 (CYP2C9, CYP2C19, CYP2E1) and dihydropyrimidine dehydrogenase (DPYD) in the Egyptian population. Br J Clin Pharmacol. 2002;53(6):596-603. [PubMed ID: 12047484].
-
24.
Adithan C, Gerard N, Vasu S, Balakrishnan R, Shashindran CH, Krishnamoorthy R. Allele and genotype frequency of CYP2C9 in Tamilnadu population. Eur J Clin Pharmacol. 2003;59(8-9):707-9. [PubMed ID: 14504850]. https://doi.org/10.1007/s00228-003-0666-3.
-
25.
Arvanitidis K, Ragia G, Iordanidou M, Kyriaki S, Xanthi A, Tavridou A, et al. Genetic polymorphisms of drug-metabolizing enzymes CYP2D6, CYP2C9, CYP2C19 and CYP3A5 in the Greek population. Fundam Clin Pharmacol. 2007;21(4):419-26. [PubMed ID: 17635181]. https://doi.org/10.1111/j.1472-8206.2007.00510.x.
-
26.
Scordo MG, Caputi AP, D'Arrigo C, Fava G, Spina E. Allele and genotype frequencies of CYP2C9, CYP2C19 and CYP2D6 in an Italian population. Pharmacol Res. 2004;50(2):195-200. [PubMed ID: 15177309]. https://doi.org/10.1016/j.phrs.2004.01.004.
-
27.
Yasar U, Eliasson E, Dahl ML, Johansson I, Ingelman-Sundberg M, Sjoqvist F. Validation of methods for CYP2C9 genotyping: frequencies of mutant alleles in a Swedish population. Biochem Biophys Res Commun. 1999;254(3):628-31. [PubMed ID: 9920790]. https://doi.org/10.1006/bbrc.1998.9992.
-
28.
Herman D, Locatelli I, Grabnar I, Peternel P, Stegnar M, Mrhar A, et al. Influence of CYP2C9 polymorphisms, demographic factors and concomitant drug therapy on warfarin metabolism and maintenance dose. Pharmacogenomics J. 2005;5(3):193-202. [PubMed ID: 15824753]. https://doi.org/10.1038/sj.tpj.6500308.
-
29.
Gaikovitch EA, Cascorbi I, Mrozikiewicz PM, Brockmoller J, Frotschl R, Kopke K, et al. Polymorphisms of drug-metabolizing enzymes CYP2C9, CYP2C19, CYP2D6, CYP1A1, NAT2 and of P-glycoprotein in a Russian population. Eur J Clin Pharmacol. 2003;59(4):303-12. [PubMed ID: 12879168]. https://doi.org/10.1007/s00228-003-0606-2.
-
30.
Huang SW, Li Q, Zhu SY, Li L, Xiong F, Jia YK, et al. SYBR Green-based real-time PCR assay for detection of VKORC1 and CYP2C9 polymorphisms that modulate warfarin dose requirement. Clin Chem Lab Med. 2009;47(1):26-31. [PubMed ID: 19117406]. https://doi.org/10.1515/CCLM.2009.008.
-
31.
Aomori T, Yamamoto K, Oguchi-Katayama A, Kawai Y, Ishidao T, Mitani Y, et al. Rapid single-nucleotide polymorphism detection of cytochrome P450 (CYP2C9) and vitamin K epoxide reductase (VKORC1) genes for the warfarin dose adjustment by the SMart-amplification process version 2. Clin Chem. 2009;55(4):804-12. [PubMed ID: 19181737]. https://doi.org/10.1373/clinchem.2008.115295.
-
32.
Kwon A, Jo SH, Im HJ, Jo YA, Park JY, Kang HJ, et al. Pharmacogenetic distribution of warfarin and its clinical significance in Korean patients during initial anticoagulation therapy. J Thromb Thrombolysis. 2011;32(4):467-73. [PubMed ID: 21713378]. https://doi.org/10.1007/s11239-011-0616-3.
-
33.
Howard R, Leathart JB, French DJ, Krishan E, Kohnke H, Wadelius M, et al. Genotyping for CYP2C9 and VKORC1 alleles by a novel point of care assay with HyBeacon(R) probes. Clin Chim Acta. 2011;412(23-24):2063-9. [PubMed ID: 21827742]. https://doi.org/10.1016/j.cca.2011.07.013.
-
34.
Esmerian MO, Mitri Z, Habbal MZ, Geryess E, Zaatari G, Alam S, et al. Influence of CYP2C9 and VKORC1 polymorphisms on warfarin and acenocoumarol in a sample of Lebanese people. J Clin Pharmacol. 2011;51(10):1418-28. [PubMed ID: 21148049]. https://doi.org/10.1177/0091270010382910.