Abstract
Background:
Computers and other electronic devices are a requisite aspect of people’s lives globally for multiple purposes, including education, entertainment, and communication.Objectives:
This study investigated the bacterial contamination of desktop computer keyboards (as a reservoir of pathogens) in different departments of Mazandaran University of Medical Sciences, Sari, Northern Iran, from September 2018 to February 2019.Methods:
In this descriptive cross-sectional study, samples were obtained from computer keyboards with sterile swabs and cultured on blood agar and eosin methylene blue agar. Standard microbiological methods were used to identify bacterial isolates. Then, the antimicrobial susceptibility testing of the isolates was performed using the Kirby-Bauer method based on the CLSI procedure.Results:
In total, 58 bacterial strains were isolated from the collected samples. The isolates included 23 (39.7%) Staphylococcus epidermidis, 15 (25.9%) Staphylococcus aureus, 14 (24.1%) Bacillus spp., 3 (5.2%) Klebsiella pneumoniae, 2 (3.4%) Pseudomonas aeruginosa, and 1 (1.7%) Escherichia coli. All Gram-positive bacteria were resistant to penicillin, whereas the most resistance rate among Gram-negative bacteria was observed against ciprofloxacin, ceftazidime, and cephalothin.Conclusions:
Due to the presence of opportunistic pathogens on computer keyboards, personal hygiene and periodic cleaning of keyboards with disinfectants is necessary to prevent the further spread of these bacteria.Keywords
Computer Keyboard Bacterial Pathogens Antibiotic Resistance Hand Hygiene
1. Background
Computers, smartphones, and laptops are electronic devices that receive information and provide results more quickly and accurately after analysis (1, 2). The bacterial contamination of computer keyboards is high due to a large number of users and their hands-on connections with the keyboards. This bacterial contamination is observed worldwide, even in developed countries (3). Pathogenic microorganisms may be present in the environments, on surfaces, and on fomites, and the hands play a major role in transmitting them (4). Many pathogenic bacteria, such as Staphylococcus aureus, coagulase-negative Staphylococci, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Escherichia coli, can survive on the keyboards for a long time, causing numerous infections (5). The keyboards always act as bacterial sources due to the frequent contact with the users’ skin (6, 7). Also, coughs and sneezes can infect computers, phones, and desktop components with various bacteria and viruses (8). Contamination of the keyboard and mouse is a potential health threat, posing a serious problem and transmitting the bacteria from person to person (9). In the United States, different bacterial strains have been isolated from computer keyboards and detected as the cause of approximately 25% of nosocomial infections (1). Antibiotic-resistant bacteria can exist on the surface of computers, especially keyboards (9).
2. Objectives
Given the key role of computer keyboards in transmitting antibiotic-resistant bacteria, we aimed to investigate the profiles and antimicrobial susceptibility patterns of bacteria isolated from computer keyboards in different sections of Mazandaran University of Medical Sciences, Sari, northern Iran.
3. Methods
3.1. Sampling and Identification Procedure
In this descriptive cross-sectional study, 58 bacterial strains were isolated from computer keyboards in the teachers’ rooms, laboratories, faculties’ information technology (IT) sites, and office rooms in educational deputies, libraries, and classrooms. The surface of computer keyboards was sampled using sterile swabs moistened with physiological saline. The swabs were then transferred to a tube containing trypticase soy broth (TSB) (Merck, Germany) and incubated for 24 h at 37°C. Then, the samples were cultured on blood agar and eosin methylene blue agar (Merck, Germany). The colonies grown on culture media were smeared and underwent Gram staining for initial bacterial identification. Then, the bacteria were identified using routine bacteriological tests, including the ability to grow on MacConkey (MAC), oxidase, catalase, lactose and glucose fermentation, citrate consumption, indole production, methyl red/Voges-Proskauer (MRVP) test, lysine decarboxylase, H2S production, coagulase, glucose fermentation, susceptibility to lysostaphin, novobiocin, and bacitracin, production of hemolysis on blood agar, growth in the presence of bile and esculin hydrolysis, and growth in the presence of 6.5% NaCl (10).
3.2. Antimicrobial Susceptibility Testing
Susceptibility patterns of the isolates were determined by the disk agar diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (11). Antibiotics used included amikacin (30 μg), gentamicin (10 μg), ciprofloxacin (5 μg), ceftazidime (30 μg), nitrofurantoin (10 μg), co-trimoxazole (25 μg), vancomycin (30 μg), erythromycin (15 μg), chloramphenicol (30 μg), penicillin (10 μg), and cephalexin (10 μg) (MAST, UK). The quality control strains for susceptibility testing were S. aureus ATCC 29213, P. aeruginosa ATCC 27853, and E. coli ATCC 25922.
4. Results
After culture and standard microbiological tests, 58 bacterial isolates were identified, including 52 (89.7%) Gram-positive and six (10.3%) Gram-negative isolates. Among the Gram-positive isolates, 23 (44.23%) were S. epidermidis, and 15 (28.84%) were S. aureus. However, among the Gram-negative isolates, there were 3 (50%) K. pneumoniae isolates and 2 (33.33%) P. aeruginosa isolates. Notably, 21 (36.2%) bacterial isolates were potential pathogens, including P. aeruginosa, K. pneumoniae, E. coli, and S. aureus, while 37 (63.79%) isolates belonged to skin flora, including S. epidermidis and Bacillus spp. (Table 1). The antibiotic susceptibility pattern of the isolated bacteria is shown in Table 2. Our findings showed that among the identified Gram-negative bacteria, all E. coli isolates were sensitive to all tested antibiotics. In this study, 100% of Gram-positive isolates were susceptible to all tested antibiotics, except chloramphenicol and nitrofurantoin. Table 3 shows the antibiotic resistance patterns of the Gram-positive bacteria isolated from the computer keyboards in different faculties. According to this table, the Gram-positive bacteria isolated from the computer keyboards of the Dental, Nursing, and Midwifery Schools were not resistant to any antibiotics except penicillin. Table 4 shows the antibiotic resistance rate of the Gram-negative bacteria isolated from computer keyboards of different schools. According to Table 5, the multi-drug resistant (MDR) phenotype was higher among Gram-negative bacteria than in Gram-positive bacteria. However, 1 (50%) isolate of P. aeruginosa was detected as MDR bacteria (resistance to at least 3 antibiotics from different classes) (12). This MDR bacillus was isolated from the keyboard of a computer located in the laboratory of the Medical School. Also, 1 (4.34%) S. epidermidis isolated from a computer in the Department of Medical Education of Allied Medical School and 1 (6.66%) S. aureus isolated from a computer in the IT Department of Health School were MDR.
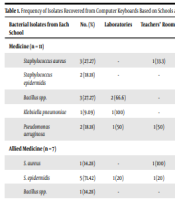
Frequency of Isolates Recovered from Computer Keyboards Based on Schools and Sources of Samples a
| Bacterial Isolates from Each School | No. (%) | Laboratories | Teachers’ Rooms | Classrooms | Office Rooms | Library | Information Technology Site |
|---|---|---|---|---|---|---|---|
| Medicine (n = 11) | |||||||
| Staphylococcus aureus | 3 (27.27) | - | 1 (33.3) | 1 (33.3) | 1 (33.3) | - | - |
| Staphylococcus epidermidis | 2 (18.18) | - | - | 1 (50) | - | 1 (50) | - |
| Bacillus spp. | 3 (27.27) | 2 (66.6) | - | - | - | - | 1 (33.3) |
| Klebsiella pneumoniae | 1 (9.09) | 1 (100) | - | - | - | - | - |
| Pseudomonas aeruginosa | 2 (18.18) | 1 (50) | 1 (50) | - | - | - | - |
| Allied Medicine (n = 7) | |||||||
| S. aureus | 1 (14.28) | - | 1 (100) | - | - | - | - |
| S. epidermidis | 5 (71.42) | 1 (20) | 1 (20) | 1 (20) | 1 (20) | - | 1 (20) |
| Bacillus spp. | 1 (14.28) | - | - | 1 (20) | - | - | - |
| Dentistry (n = 8) | |||||||
| S. aureus | 3 (37.5) | - | - | 1 (33.3) | 1 (33.3) | - | 1 (33.3) |
| S. epidermidis | 4 (50) | 1 (25) | 2 (50) | - | - | 1 (25) | - |
| Bacillus spp. | 1 (12.5) | - | - | 1 (100) | - | - | - |
| Pharmacy (n = 9) | |||||||
| S. aureus | 3 (33.33) | - | - | 1 (33.3) | 1 (33.3) | 1 (33.3) | |
| S. epidermidis | 2 (22.22) | - | - | 1 (50) | - | - | 1 (50) |
| Bacillus spp. | 3 (33.33) | 1 (33.3) | 2 (66.7) | ||||
| K. pneumoniae | 1 (11.11) | 1 (100) | - | - | - | - | - |
| Health (n = 9) | |||||||
| S. aureus | 5 (55.55) | 1 (20) | 1 (20) | - | 1 (20) | 1 (20) | 1 (20) |
| S. epidermidis | 2 (22.22) | - | 1 (50) | - | - | 1 (50) | - |
| Bacillus spp. | 2 (22.22) | - | 2 (100) | - | - | - | |
| Nursing (n = 7) | |||||||
| S. epidermidis | 3 (42.85) | 1 (33.3) | 2 (66.7) | - | - | - | - |
| Bacillus spp. | 2 (28.57) | - | - | 1 (50) | 1 (50) | - | - |
| K. pneumonia | 1 (14.28) | - | - | - | - | - | 1 (100) |
| Escherichia coli | 1 (14.28) | - | - | 1 (100) | - | - | - |
| Midwifery (n = 7) | |||||||
| S. epidermidis | 5 (71.42) | - | 2 (40) | 2 (40) | - | - | 1 (20) |
| Bacillus spp. | 2 (28.57) | - | - | - | 1 (50) | 1 (50) | - |
| Total (58) | 10 | 14 | 14 | 7 | 6 | 7 |
Antibiotic Resistance Pattern of Gram-Positive and Gram-Negative Bacteria Isolated from Computer Keyboards
| Antibiotics and Classes | No. (%) of Resistance Rate Among Different Bacterial Isolates | |||||
|---|---|---|---|---|---|---|
| Staphylococcus aureus (n = 15) | Staphylococcus epidermidis (n = 23) | Bacillus spp. (n = 14) | Klebsiella pneumoniae (n = 3) | Pseudomonas aeruginosa (n = 2) | Escherichia coli (n = 1) | |
| Nitrofurantoin | ||||||
| R | 2 (13.3) | 1 (4.3) | 1 (7.1) | - | - | - |
| S | 16 (86.7) | 22 (95.7) | 12 (95.7) | 3 (100) | 2 (100) | 1 (100) |
| I | - | - | 1 (7.1) | - | - | - |
| Chloramphenicol | ||||||
| R | 1 (6.7) | 1 (4.3) | 1 (7.1) | - | - | - |
| S | 14 (93.3) | 22 (95.7) | 13 (92.9) | 3 (100) | 2 (100) | 1 (100) |
| Gentamicin | ||||||
| R | - | - | - | - | - | - |
| S | 15 (100) | 23 (100) | 14 (100) | 3 (100) | 2 (100) | 1 (100) |
| Vancomycin | ||||||
| R | - | - | - | - | - | - |
| S | 15 (100) | 23 (100) | 14 (100) | - | - | - |
| Penicillin | ||||||
| R | 14 (93.3) | 22 (95.7) | 10 (71.4) | - | - | - |
| S | 1 (6.7) | 1 (4.3) | 4 (28.6) | - | - | - |
| Amikacin | ||||||
| R | - | - | - | - | - | - |
| S | - | - | - | 3 (100) | 2 (100) | 1 (100) |
| Ceftazidime | ||||||
| R | - | - | - | 1 (33.3) | 2 (100) | - |
| S | - | - | - | 2 (66.7) | - | 1 (100) |
| Ciprofloxacin | ||||||
| R | - | - | - | - | 2 (100) | - |
| S | 15 (100) | 23 (100) | 14 (100) | 3 (100) | - | 1 (100) |
| Co-trimoxazole | ||||||
| R | - | - | - | - | - | - |
| S | 15 (100) | 23 (100) | 14 (100) | 3 (100) | 2 (100) | 1 (100) |
| Cephalothin | ||||||
| R | - | - | - | 1 (33.3) | 1 (50) | - |
| S | 15 (100) | 23 (100) | 14 (100) | 2 (66.7) | 1 (50) | 1 (100) |
| Erythromycin | ||||||
| R | - | - | - | - | - | - |
| S | 15 (100) | 23 (100) | 14 (100) | - | - | - |
Antibiotic Resistance Pattern of Gram-Positive Bacteria from Different Faculties
| Antibiotics and Classes | No. (%) of Resistance and Susceptibility Rate of Gram-Positive Bacteria Isolated from the School of | ||||||
|---|---|---|---|---|---|---|---|
| Medicine (n = 8) | Allied Medicine (n = 7) | Dentistry (n = 8) | Pharmacy (n = 8) | Health (n = 9) | Nursing (n = 5) | Midwifery (n = 7) | |
| Nitrofurantoin | |||||||
| R | 1 (12.5) | 1 (14.3) | - | 1 (12.5) | 1 (11.1) | - | - |
| S | 7 (87.5) | 6 (85.7) | 8 (100) | 7 (87.5) | 7 (77.8) | 5 (100) | 7 (100) |
| I | - | - | - | - | 1 (11.1) | - | - |
| Chloramphenicol | |||||||
| R | - | 1 (14.3) | - | 1 (12.5) | 1 (11.1) | - | - |
| S | 8 (100) | 6 (85.7) | 8 (100) | 7 (87.5) | 8 (88.9) | 5 (100) | 7 (100) |
| Gentamicin | |||||||
| R | - | - | - | - | - | - | - |
| S | 8 (100) | 7 (100) | 8 (100) | 8 (100) | 9 (100) | 5 (100) | 7 (100) |
| Vancomycin | |||||||
| R | - | - | - | - | - | - | - |
| S | 8 (100) | 7 (100) | 8 (100) | 8 (100) | 9 (100) | 5 (100) | 7 (100) |
| Penicillin | |||||||
| R | 6 (75) | 7 (100) | 8 (100) | 5 (62.5) | 8 (88.9) | 5 (100) | 7 (100) |
| S | 2 (25) | - | - | 3 (37.5) | 1 (11.1) | - | - |
| Ciprofloxacin | |||||||
| R | - | - | - | - | - | - | - |
| S | 8 (100) | 7 (100) | 8 (100) | 8 (100) | 9 (100) | 5 (100) | 7 (100) |
| Co-trimoxazole | |||||||
| R | - | - | - | - | - | - | - |
| S | 8 (100) | 7 (100) | 8 (100) | 8 (100) | 9 (100) | 5 (100) | 7 (100) |
| Cephalothin | |||||||
| R | - | - | - | - | - | - | - |
| S | 8 (100) | 7 (100) | 8 (100) | 8 (100) | 9 (100) | 5 (100) | 7 (100) |
| Erythromycin | |||||||
| R | - | - | - | - | - | - | - |
| S | 8 (100) | 7 (100) | 8 (100) | 8 (100) | 9 (100) | 5 (100) | 7 (100) |
Antibiotic Resistance Pattern of Gram-Negative Bacteria in Different Faculties
| Antibiotics and Classified | No. (%) of Resistance and Susceptibility Rate of Gram-Negative Bacteria Isolated from the School of | ||||||
|---|---|---|---|---|---|---|---|
| Medicine (n = 3) | Allied Medicine (n = 0) | Dentistry (n = 0) | Pharmacy (n = 1) | Health (n = 0) | Nursing (n = 2) | Midwifery (n = 0) | |
| Nitrofurantoin | |||||||
| R | - | - | - | - | - | - | - |
| S | 3 (100) | 1 (100) | 2 (100) | - | |||
| Chloramphenicol | |||||||
| R | 3 (100) | - | - | 1 (100) | 2 (100) | - | |
| S | - | - | - | - | - | - | - |
| Gentamicin | |||||||
| R | - | - | - | - | - | - | - |
| S | 3 (100) | - | - | 1 (100) | - | 2 (100) | - |
| Amikacin | |||||||
| R | - | - | - | - | - | - | |
| S | 3 (100) | - | - | 1 (100) | - | 2 (100) | - |
| Ceftazidime | |||||||
| R | 3 (100) | - | - | - | - | - | |
| S | - | - | - | 1 (100) | - | 2 (100) | - |
| Ciprofloxacin | |||||||
| R | 2 (66.7) | - | - | - | - | - | |
| S | 1 (33.3) | - | - | 1 (100) | - | 2 (100) | - |
| Co-trimoxazole | |||||||
| R | - | - | - | - | - | - | |
| S | 3 (100) | - | - | 1 (100) | - | 1 (100) | - |
| Cephalothin | |||||||
| R | 2 (66.7) | - | - | - | - | - | |
| S | 1 (33.3) | - | - | 1 (100) | - | 2 (100) | - |
Prevalence of Resistance to Several Antibiotics Among Bacterial Isolates
| Bacteria | No. (%) of Isolates Resistant to | ||||
|---|---|---|---|---|---|
| No Drug | 1 Drug | 2 Drugs | 3 Drugs | 4 Drugs | |
| Staphylococcus epidermidis (n = 23) | 1 (4.34) | 21 (91.30) | - | 1 (4.34) | - |
| Staphylococcus aureus (n = 15) | - | 14 (93.33) | - | 1 (6.66) | - |
| Bacillus spp. (n = 14) | 3 (21.42) | 10 (71.42) | 1 (7.14) | - | - |
| Klebsiella pneumonia (n = 3) | 2 (66.66) | - | 1 (33.33) | - | - |
| Pseudomonas aeruginosa (n = 2) | - | - | 1 (100) | 1 (100) | - |
| Escherichia coli (n = 1) | - | - | - | - | - |
5. Discussion
The results showed that S. epidermidis, Bacillus species, and S. aureus were the most common bacterial agents contaminating computer keyboards. These results are consistent with other studies in our region and neighboring and non-neighboring countries (3, 13, 14). In a study conducted by Awe et al. at Salem University, Lokoja, Nigeria, similar to our research, they found numerous microorganisms on the keyboard and mouse of the computers in the university library (15). However, in our study, no bacteria were observed in the library of the Allied Medical School. According to our review, the reason was the frequent use of disinfectants for book surfaces and shelves in the library. Disinfectants play an important role in the control of bacteria, as other studies on microbial contamination of computer keyboards and mobile handsets have confirmed the role of disinfectants such as ethanol in reducing and even killing bacteria (1, 16, 17). Also, the Medical School Laboratory showed the highest prevalence of Gram-positive/negative bacteria in the present study, so that even P. aeruginosa, as an opportunistic pathogen, was isolated from the computers of this laboratory. This may be due to the use of clinical samples sent to the laboratory for research projects and training of medical students. The prevalence of Bacillus spp. in this study was also consistent with other studies (3, 13). This Gram-positive bacterium is found as saprophytes in soil, water, and air and is transmitted by hand (18). Besides, S. aureus is a natural skin and nose flora that causes numerous infections. In our study, 25.9% of the identified isolates were S. aureus. Two other studies from Nigeria and Iran also reported 39% and 23% contamination rates of computer keyboards with S. aureus, respectively (16, 19). They also pointed out that the computer emits radiation that is effective on microorganisms and that the abundance of S. aureus on the computer keyboard could be due to its high tolerance against the radiation emitted by the computer (20). A study by Mehdinejad et al. from Iran reported Bacillus spp., coagulase-negative Staphylococci, S. aureus, and Enterobacteriaceae, similar to our study, although their isolates were collected just from the School of Medicine (13). This outbreak could be due to the close contact of medical students with patients in educational hospitals and the possibility of bacterial transmission to computer keyboards. Bacterial contamination of the keyboards has posed a threat to public health so that according to researchers, the bacteria grown on keyboards of the computers are more dangerous for human health (2). In another survey from Sudan, coagulase-negative Staphylococci were isolated in 56% of cases, which was higher than our study (21). It should be noted that their isolates were collected from the elevator keypad, while these devices are more contaminated due to wider and frequent skin contact. When sampling is similar, the type and prevalence of the bacteria are usually somewhat similar. However, the prevalence of S. epidermidis, Bacillus spp., S. aureus, and P. aeruginosa in our study is very close to research performed by Alemu et al. in Ethiopia (3).
On the other hand, in a study conducted in a dental educational institution in Mashhad, northeastern Iran, only normal flora bacteria were isolated from the control population of non-medical persons (22). In addition, the computers in the classrooms and the teachers’ rooms were the most contaminated computers in this study. The presence of bacteria such as S. aureus, P. aeruginosa, and E. coli on the keyboards of these computers could be due to the high level of communication between professors and laboratories and the possibility of bacterial transfer to the computers of their rooms and classrooms. This result demonstrates the need for frequent and proper hand disinfection of professors; also, because 1 of the P. aeruginosa isolates was MDR, it may be a serious health hazard. In the present study, all Gram-negative isolated bacteria were susceptible to amikacin and gentamicin, consistent with the study by Mohammed (23). Also, Gram-negative bacteria showed no resistance to nitrofurantoin, chloramphenicol, gentamicin, amikacin, and co-trimoxazole. These results suggest that environmentally sensitive strains may have contributed to these infections. As an argument, the Gram-negative bacteria isolated from the keyboards of the computers in our study, except in the Medical School, were resistant only to chloramphenicol, and 100% of the isolates were sensitive to the rest of the tested antibiotics. The only concern of the present study was about P. aeruginosa, where both isolates were resistant to ceftazidime and ciprofloxacin, 1 of which was MDR.
On the other hand, Gram-negative bacteria isolated from the Medical School’s computer keyboards showed significant resistance rates to chloramphenicol, ciprofloxacin, ceftazidime, and cephalothin. Furthermore, multiple drug resistance phenotypes and antibiotic resistance rates were higher in Gram-negative than in Gram-positive bacteria, consistent with an earlier study (3). Also, in this study, the highest antibiotic resistance was observed among Gram-positive isolates against penicillin, while no resistance was found to gentamicin, vancomycin, co-trimoxazole, cephalothin, and erythromycin, which was concordant with a study in Ethiopia (3). We observed that Gram-positive bacteria isolated from the keyboards of computers in Dental, Nursing, and Midwifery Schools showed no resistance to antibiotics, except penicillin. The higher resistance to penicillin was probably because most Gram-positive isolates (73.07%) were Staphylococci, which are no longer sensitive to penicillin due to the high production of beta-lactamases (24). Moreover, 1 (4.34%) isolate of S. epidermidis, 1 (6.66%) isolate of S. aureus, and 1 (7.14%) isolate of Bacillus spp. were MDR.
Regarding the presence of Bacillus spp. on inanimate surfaces, the prevalence in the previous study by Sedighi et al. was lower than in our study, whereas they evaluated only cell phones (25). This may be because the cell phone is a personal device, so people feel more responsible for cleaning it. Considering the results of the current study and other studies, as well as the worrisome prevalence of coronavirus in the current period, we suggest the observance of personal hygiene and the use of disinfectants such as quaternary ammonium compounds to clean computer keyboards and other inanimate surfaces and the hands of users who are more connected with these surfaces.
References
-
1.
Sebre S, Abegaz WE, Seman A, Awoke T, Desalegn Z, Mihret W, et al. Bacterial Profiles and Antimicrobial Susceptibility Pattern of Isolates from Inanimate Hospital Environments at Tikur Anbessa Specialized Teaching Hospital, Addis Ababa, Ethiopia. Infect Drug Resist. 2020;13:4439-48. [PubMed ID: 33364791]. [PubMed Central ID: PMC7751703]. https://doi.org/10.2147/IDR.S286293.
-
2.
Olu-Taiwo M, Laryea CA, Kweku Mykels D, Forson AO. Multidrug-Resistant Bacteria on the Mobile Phones and Computer Keyboards of Healthcare University Students in Ghana. Can J Infect Dis Med Microbiol. 2021;2021:6647959. [PubMed ID: 33936348]. [PubMed Central ID: PMC8062198]. https://doi.org/10.1155/2021/6647959.
-
3.
Alemu A, Misganaw D, Wondimeneh Y. Bacterial profile and their antimicrobial susceptibility patterns of computer keyboards and mice at Gondar University Hospital, Northwest Ethiopia. Biomed Biotechnol. 2015;3(1):1-7.
-
4.
Gerba CP. Environmentally Transmitted Pathogens. In: Maier RM, Pepper IL, Gerba CP, editors. Environmental Microbiology. 2nd ed. Massachusetts, USA: Academic Press; 2009. p. 445-84. https://doi.org/10.1016/b978-0-12-370519-8.00022-5.
-
5.
Enemuor SC, Apeh T, Oguntibeju OO. Microorganisms associated with computer keyboards and mice in a university environment. Afr J Microbiol Res. 2012;6(20). https://doi.org/10.5897/ajmr11.1288.
-
6.
Ukamaka Umeanaeto P, Chukwuma Okafor U, Chisom Unam M, Chidimma Ilo C, Chigozie Okoli C, Chinyere Afulukwe S, et al. Assessment of Parasites and Bacterial Contamination of Office Door Handles in Nnamdi Azikiwe University, Awka, Anambra State. American Journal of Biomedical and Life Sciences. 2021;9(2). https://doi.org/10.11648/j.ajbls.20210902.13.
-
7.
Mbithi JN, Springthorpe VS, Boulet JR, Sattar SA. Survival of hepatitis A virus on human hands and its transfer on contact with animate and inanimate surfaces. J Clin Microbiol. 1992;30(4):757-63. [PubMed ID: 1315331]. [PubMed Central ID: PMC265157]. https://doi.org/10.1128/jcm.30.4.757-763.1992.
-
8.
Lopez GU, Gerba CP, Tamimi AH, Kitajima M, Maxwell SL, Rose JB. Transfer efficiency of bacteria and viruses from porous and nonporous fomites to fingers under different relative humidity conditions. Appl Environ Microbiol. 2013;79(18):5728-34. [PubMed ID: 23851098]. [PubMed Central ID: PMC3754157]. https://doi.org/10.1128/AEM.01030-13.
-
9.
Koscova J, Hurnikova Z, Pistl J. Degree of Bacterial Contamination of Mobile Phone and Computer Keyboard Surfaces and Efficacy of Disinfection with Chlorhexidine Digluconate and Triclosan to Its Reduction. Int J Environ Res Public Health. 2018;15(10). [PubMed ID: 30322055]. [PubMed Central ID: PMC6210060]. https://doi.org/10.3390/ijerph15102238.
-
10.
Mahon CR, Lehman DC, Manuselis G. Textbook of diagnostic microbiology-e-book. Amsterdam, the Netherlands: Elsevier Health Sciences; 2018.
-
11.
Humphries RM, Kircher S, Ferrell A, Krause KM, Malherbe R, Hsiung A, et al. The Continued Value of Disk Diffusion for Assessing Antimicrobial Susceptibility in Clinical Laboratories: Report from the Clinical and Laboratory Standards Institute Methods Development and Standardization Working Group. J Clin Microbiol. 2018;56(8). [PubMed ID: 29743302]. [PubMed Central ID: PMC6062797]. https://doi.org/10.1128/JCM.00437-18.
-
12.
Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268-81. [PubMed ID: 21793988]. https://doi.org/10.1111/j.1469-0691.2011.03570.x.
-
13.
Mehdinejad M, Khosravi AD, Afzali M, Mahmoudabadi AZ. Study of bacterial contamination of keyboard and mouse in a medical school computer center. HealthMED. 2012;6(3):889-92.
-
14.
Al Ghamdi AK, Abdelmalek SM, Ashshi AM, Faidah H, Shukri H, Jiman Fatani AA. Bacterial contamination of computer keyboards and mice, elevator buttons and shopping carts. Afr J Microbiol Res. 2011;5(23):3998-4003. https://doi.org/10.5897/ajmr11.770.
-
15.
Awe S, Eniola K, Livingstone S. Bacteriological assessment of computer keyboards and mouse used in Salem University, Lokoja. Am J Res Commun. 2013;1(12):398-412.
-
16.
Karbasizade V, Sichani M, Parsafar S. Bacterial contamination of computer keyboards in hospitals in Isfahan in Iran. Int J Biosci. 2014:320-4. https://doi.org/10.12692/ijb/4.1.320-324.
-
17.
Messina G, Ceriale E, Burgassi S, Russo C, Defranceschi C, Mariani L, et al. Impact of a disinfecting technique on microbial contamination of computer keyboards and telephone handsets. J Hosp Adm. 2013;2(4). https://doi.org/10.5430/jha.v2n4p1.
-
18.
Ehling-Schulz M, Lereclus D, Koehler TM. The Bacillus cereus Group: Bacillus Species with Pathogenic Potential. Microbiol Spectr. 2019;7(3). [PubMed ID: 31111815]. [PubMed Central ID: PMC6530592]. https://doi.org/10.1128/microbiolspec.GPP3-0032-2018.
-
19.
Muhammad RH, Yaro CA, Balarabe Musa B, Safiyya SB. Isolation and Identification of Bacteria Associated with Computer Keyboards and Mouse at Various Business Centres in Dutse Metropolis Jigawa State, Nigeria. Int J Curr Microbiol App Sci. 2016;5(8):811-7. https://doi.org/10.20546/ijcmas.2016.508.090.
-
20.
Rapacka-Zdonczyk A, Wozniak A, Pieranski M, Woziwodzka A, Bielawski KP, Grinholc M. Development of Staphylococcus aureus tolerance to antimicrobial photodynamic inactivation and antimicrobial blue light upon sub-lethal treatment. Sci Rep. 2019;9(1):9423. [PubMed ID: 31263139]. [PubMed Central ID: PMC6603016]. https://doi.org/10.1038/s41598-019-45962-x.
-
21.
Kandel CE, Simor AE, Redelmeier DA. Elevator buttons as unrecognized sources of bacterial colonization in hospitals. Open Med. 2014;8(3):e81-6. [PubMed ID: 25426176]. [PubMed Central ID: PMC4242253].
-
22.
Movahhed T, Dehghani M, Ghoddusi T. Evaluation of microbial contamination of mobile phones and computer mice and keyboards in a dental school. J Dent Mater Tech. 2018;7(2):78-82.
-
23.
Mohammed SSE. Detection of Multi-drugs Resistance among Bacteria Isolated from Computers’ Keyboards. Khartoumm Sudan: Sudan University of Science & Technology; 2015.
-
24.
Kesharwani AK, Mishra J. Detection of β-lactamase and antibiotic susceptibility of clinical isolates of Staphylococcus aureus. Biocatal Agric Biotechnol. 2019;17:720-5. https://doi.org/10.1016/j.bcab.2018.12.012.
-
25.
Sedighi I, Alikhani MY, Ramezani S, Nazari M, Mozaffari Nejad AS. Bacterial Contamination of Mobile Phones of Health Care Providers in a Teaching Hospital in Hamadan Province, Iran. Arch Clin Infect Dis. 2015;10(2). https://doi.org/10.5812/archcid.10(2)2015.22104.