Abstract
Keywords
1. Background
Chlamydia psittaci (C. psittaci), the major causative agent of psittacosis, is a widespread, Gram-negative, intracellular bacterium, which most frequently infects Psittaciforms; however; it can also infect numerous other avian species such as pigeons and sparrows and mammalian species including humans (1, 2). C. psittaci can infect humans via the inhalation of aerosols, feather particles or dried respiratory tract secretions from diseased or subclinically infected birds (3, 4). Human-to-human transmission is rare, but may occur (5). Human infection with C. psittaci can cause psittacosis that is occasionally associated with respiratory disease; however, clinical complications such as malaise, myalgia, and multi-organ failure may also occur; occasionally, leading to fatal outcomes in untreated patients (3, 4, 6). Factors such as the chlamydial strain, immunity status of the host, concurrent or secondary infections, as well as therapeutic management, can influence the severity of the clinical signs (7, 8). Moreover, no vaccines or valid agents are available against C. psittaci, which makes it more complicated to deal with (7, 9, 10).
In birds, C. psittaci might manifest itself as an upper respiratory infection that is characterized by nasal and ocular discharge, diarrhea, anorexia, respiratory distress, sinusitis, rhinitis and conjunctivitis (10). C. psittaci has been isolated from a variety of wild bird species, particularly Columbiformes such as collared doves, urban pigeons, and occasionally passerines (11). In a comprehensive literature search of epidemiological studies and reports of transmissions of disease from feral pigeons to humans by Haag-Wackernagel and Moch. They showed that feral pigeons harbored 60 different human pathogenic organisms, but only seven were transmitted to humans, and most of the commonly transmitted pathogens continue to be C. psittaci and Cryptococcus neoformans (12). In the past decade, severe cases of human psittacosis have been reported including some cases from Asia (13-15). The potential of pigeons (Columba livia domestica) as a source of human psittacosis is supported worldwide by relatively high seroconversion rates in the birds (16).
Currently, C. psittaci is classified into ten genotypes of A-G and E/B (avian species) and M56 and WC (mammals) based on sequencing the major outer-membrane protein gene (ompA), each genotype displaying overlapping host preferences and virulence characteristics (17-19). All the C. psittaci genotypes, including the avian genotypes, particularly A, B, and E/B, should be considered potential zoonotic (20, 21). The C. psittaci infecting wild passerines mainly belongs to the genotypes A and B (18, 22, 23), with a low fatality rate in the affected birds (1). Although pigeons harbor a variety of C. psittaci genotypes, the most common genotypes in pigeons are B and E (24).
2. Objectives
This study aimed at evaluating the occurrence and prevalence of C. psittaci in pigeons, passerines, as well as the possible zoonotic transmission to in-contact humans in Chaharmahal-va-Bakhtiari Province, Iran. The findings of this study will provide new insights into the prevalence and genetic diversity of C. psittaci in pigeons, passerines, and humans and its zoonotic potential.
3. Methods
3.1. Study Area
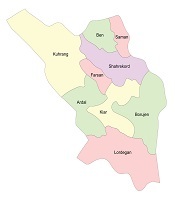
This study was performed in six different origins in Chaharmahal-va-Bakhtiari Province, a province located in the southwestern part of Iran (32° 32’ N longitude-50°85’ E latitude) (Figure 1). It covers an area of 16,332 km2, with a population of approximately 947,763 (2016 census). Shahrekord, the capital city of this province, is Iran’s highest city in altitude at 2,070 m above the sea level and is known as the Roof of Iran. The annual average temperature in Shahrekord is about 11°C but the minimum and maximum absolute temperatures recorded in Shahrekord during the last 30 years have been -32°C and 42°C, respectively.
Map of Iran and the location of Chaharmahal-va-Bakhtiari Province, where the samples were collected. The graphs are shown prevalence of C. psittaci in six locations of Chaharmahal-va-Bakhtiari Province.
3.2. Sampling
A total of 75 specimens of urban pigeon, 75 specimens of the house sparrow, and 30 pharyngeal swabs of individuals who worked in pet-markets were collected from six different zones of Chaharmal-va-Bakhtiari (Figure 1). The free-living birds were attracted using food baits and a special net was applied for their trapping. Fresh cloacal swabs were sampled immediately, and the birds were released afterward. All birds appeared to be healthy in terms of body conditions, as confirmed by a veterinarian.
3.3. DNA Extraction and PCR
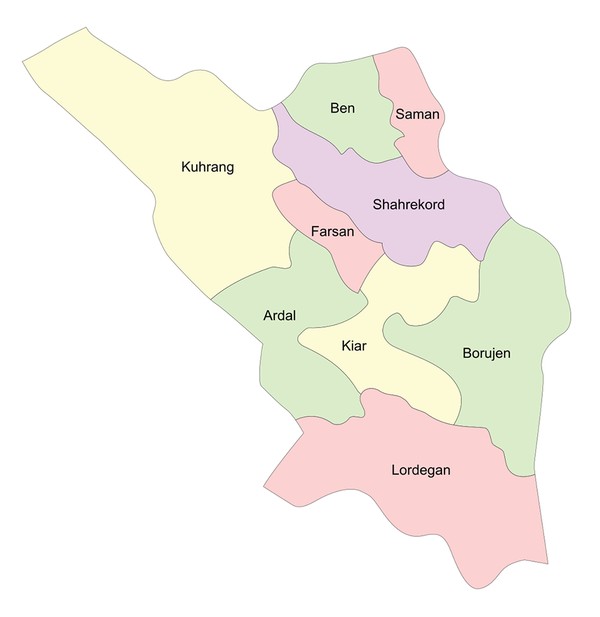
Genomic DNA extraction was performed using the DNPTM DNA extraction Kit (SinaClon, Karaj, Iran) according to the manufacturer’s instructions. C. psittaci PCR was performed using a previously described protocol 25 and a PCR method developed in this study. The two C. psittaci PCR procedures target an 82-bp (primers: CpsittF and CPsittR) and a 157-bp fragment of the OmpA gene (primers: Cp.sG1F and Cp.sG1R) (Table 1 and Figure 2).
PCR Primers Used in This Study
| Primer Name | Primer Sequence 5’ to 3’ | Amplicon Size, bp | Reference |
|---|---|---|---|
| Cpsitt | F: CGCTCTCTCCTTACAAGCC | 82 | (19) |
| R: AGCACCTTCCCACATAGTG | |||
| CPsittGeno | F: GCTACGGGTTCCGCTCT | 1,041 | (19) |
| R: TTTGTTGATYTGAATCGAAGC | |||
| CPsitt | Finner: CGCTCTCTCCTTACAAGCC | 1024 | (19) |
| Rinner: GATCTGAATCGAAGCAATTTG | |||
| Cp.sG1 | F: GCCTTAAACATTTGGGATCG | 157 | The present study |
| R: CGTTAGGAAGTTGCATTGGA | |||
| Cp.sG2 | F: GACACTCCTCAAAGCCATTA | 1024 | The present study |
| R: CGCCAATATATGGAACAAGC | |||
| Cp.sG3 | F: TGCAACTTTAGGAGCTGA | 622 | The present study |
| R: GTTCTGATAGCGGGACAA |
Gel electrophoresis of PCR amplicons of DNA samples. A, Lane 1, 50 bp DNA ladder, (SinaClon, Karaj, Iran); lane 2, negative control; lane 3 - 7 specific bands of C. psittaci DNA, 157 bp, 505 bp (primers; Cp.sG1 F and Cp.sG2 R), 622 bp, 1024 bp (primers; Cp.sG2 F and Cp.sG2R) and 1389 bp (primers; Cp.sG2 F and Cp.sG3R), respectively; B, lane 1, 50 bp DNA ladder, (SinaClon, Karaj, Iran); lane 2, negative control; lane 3 - 5 specific bands of C. psittaci DNA, 82 bp, 1024 bp (primers; CPsittFinner and CPsittRinner) and 1041 bp, respectively.
3.4. PCR Reaction Condition
The PCR ingredients (25 µL) consisted of 12.5 µL of 2× PCR Master mix (SinaClon, Karaj, Iran), a 0.3 M concentration of each primer, 3 µL of extracted DNA and water. PCR reactions were carried out using an Applied Biosystems 2720 Thermal Cycler (Applied Biosystems, Foster City, CA, USA) under the following conditions: an initial denaturation temperature of 95°C for 5 min followed by 45 cycles of 94°C for 15 s, 57°C for 35 s and 72°C for 45 s. Five microliters of the PCR products were analyzed by 1% agarose gel electrophoresis and UV imaging system (UVItec, Cambridge, United Kingdom) (Figure 2).
3.5. Genotyping
PCR-positive samples were genotyped by OmpA sequence analysis as previously described (25). In brief, a part of the OmpA gene was amplified with the primer CPsittGeno that resulted in a 1041-bp amplicon (25) and the overlapping sequences were obtained with following sequencing primers: CPsittFinner and CPsittRinner (yielding an amplicon with a size of 1024 bp) (25), Cp.sG1 F and Cp.sG2 R (amplicon size: 505 bp), Cp.sG2 F and Cp.sG2R (amplicon size: 1024 bp) and Cp.sG2 F and Cp.sG3R (amplicon size: 1389 bp) (Table 1 and Figure 2). PCR-positive samples were also genotyped by Cps.GD2 and Cps.GD3 primers (Table 1). These primers target the conserved regions of the OmpA gene often used for the genotyping of C. psittaci in the under-study birds.
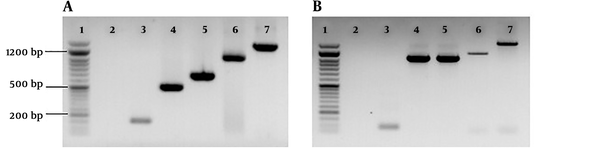
To obtain sufficient products for sequence analysis, the positive amplicons of the expected sizes were reamplified. The reaction mixture for reamplification (50 µL) included 2 µL of PCR product, 25 µL of 2× PCR Master mix (SinaClon, Karaj, Iran), 0.2 M concentration of each primer and water. The PCR conditions were as outlined above. Then, the PCR positive samples were sequenced by Macrogen (Seoul, South Korea). The resultant sequence reads were examined for ambiguity and trimmed to remove low quality ends using Chromatogram Explorer Lite version 3 (Heracle BioSoft S.R.L., Pitesti, Romania). Sequence alignment was done using the program DNA Baser Sequence Assembler version 4 (Heracle BioSoft S.R.L., Pitesti, Romania). Other sequences used in the alignment were 9 reference sequences for the C. psittaciOmpA genotypes and the C. caviae strain (AF269282). Reference sequences of the nine C. psittaci genotypes available at the database of GenBank, including A (accession number AY762608), B (AF269265), C (L25436), D (AF269266), E (X12647), F (AF269259), E/B (AY762613), M56 (AF269268), and WC (AF269269), were analyzed (Figure 3). The phylogenetic tree was constructed by Neighbor-Joining method (26) implemented in the MEGA 6 software (27). The bootstrap values were calculated for 2,000 replicates of the alignment (28). The sequences of C. psittaci strains (Sku/Pi2, Sku/Pi21, Sku/Pi22, CpsiSku/Pi64, Sku/Pas21, Sku/Pas43, CpsiSku/Pas49) detected in this study were submitted to the GenBank and assigned the accession numbers KM609420, KM609421, KM609422, MG323309, KM609418, KM609419, and MG323308, respectively.
Neighbor-joining (NJ) phylogenetic tree based on the comparison of outer membrane protein A (OmpA) gene fragments of C. psittaci strains with reference sequences in this study. The sequences detected in this study are shown with black field rhombus. The tree was rooted with the C. caviae ompA sequence. Bootstrap values are shown if the reliability is equal to or greater than 50%. The Genotype/Strain name is followed by the accession number for each branch of the tree (Strain name-Genotype). All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA6.
4. Results
A total of 75 urban pigeon specimens, 75 house sparrow specimens, and 30 human pharyngeal swab samples were examined using PCR specific for C. psittaci OmpA gene. From the 75 pigeon samples tested, 19 (25.3%) were positive for the C. psittaci DNA. The number of positive passerine samples for C. psittaci was 14 out of 75 (14.75%). These PCR-positive samples were further characterized by OmpA genotyping. The homologous sequences were excluded from the comparison, and another sequence was submitted to GenBank. The representative sequences of C. psittaci were downloaded from the GenBank database. Then, a dataset for the analysis of the OmpA gene was composed of the reference sequences and the resulting sequences. The OmpA DNA sequences of the 19 PCR positive pigeon samples grouped all the C. psittaci strains as genotype B. The OmpA sequences of the pigeon strains were highly homologous, a similarity of more than 97.4 % (Figure 4). These genotype B strains revealed 99.6% to 100% similarities to the reference B genotype (accession number AY762609.1). The nucleotide sequences of the 14 passerine C. psittaci strains clustered the strains with those of genotype A. The sequences had the highest similarity to each other, more than 99.5% and to those of reference A strain (accession number AY762608.1) with more than 98.8% similarity (Figure 4). The nucleotide sequences of the Iranian C. psittaci strains were highly homologous with an average nucleotide similarity of 97.2%. The three PCR positive human pharyngeal samples only produced 82 bp and 157 bp DNA sequences, both identical to those of human genotype B reference strains 41A12 (accession number AY762609.1).
Percentage identity matrix of detected sequences (representative sequences) and reference sequences of C. psittaci and C. cavae of OmpA gene

5. Discussion
Chlamydia psittaci is an avian pathogen and is generally prevalent in wild and companion birds as well as in poultry. However, it can also cause zoonotic disease in humans. The purpose of this study was to survey and collect baseline data in Chaharmahal-va-Bakhtiari Province of Iran on the prevalence of C. psittaci in pigeons (Columba livia) and passerines (Passer domesticus). In addition, pharyngeal swabs from pet-shop workers in these areas were also analyzed for the presence and the type of C. psittaci.
PCR-based methods are highly sensitive and can detect very low copy numbers of chlamydial DNA in samples along with providing information on their genetic diversity and assessing their potential zoonosis (17, 20). As there is no universally agreed PCR protocol for the detection of chlamydial DNA, various techniques and procedures, differing in sample type and size and in the targeted genes and primers, are employed (17, 18, 25, 29, 30). Therefore, different results have been achieved or in some studies genotyping of positive cases failed for various reasons such as the presence of only small amounts of DNA or due to the presence of unknown OmpA genotypes (22, 30).
Avian species harbor a number of zoonotic pathogens, and those living in close proximity to humans, particularly in public places, can be considered potential public health threats. This study shows that 25.3% of urban pigeon samples and 18.6% of passerine samples in Chahrmahal-va-Bakhtiari harbor C. psittaci. Genotype B was found in pigeon samples, and genotype A was found in passerine samples. Our results show that C. psittaci is highly prevalent among the urban pigeons and passerines. Three out of 30 (10 %) human pharyngeal swabs also were positive for C. psittaci DNA, indicating the possibility of lateral transmission to those people who often come in close contact with these avian species. We were unable to genotype human PCR-positive samples.
The findings of the present study are in accordance with those of other studies. Three hundred and twelve fecal samples from 18 bird species of wild passerine were examined by Olsen et al. (31), using PCR and DNA sequencing. C. psittaci DNA was demonstrated in 9 (2.9 %) birds indicating wild passerine birds as carriers of C. psittaci. C. psittaci genotype was not determined in the study. In a study conducted in Amsterdam, from the 331 feral pigeons examined, 26 (7.9 %) were positive for C. psittaci and 10 were typed as genotype B (25). A C. psittaci prevalence of 16.8 % (40/238) in pigeons is also reported from Sao Paulo State, Brazil (32). Salinas et al. (33) reported one of the largest series on the prevalence of C. psittaci in feral pigeons. In their study, C. psittaci was found by culture in 18% (7/39; 95% CI, 9% to 33%) of fecal samples, a prevalence that is lower than our results obtained by PCR. In a study by Feng et al. (9), C. psittaci was detected in 27 out of 136 fresh fecal samples (19.9%) in psittacine birds using TaqMan MGB real-time PCR, which is a highly sensitive and specific technique for detecting chlamydial DNA.
In a study conducted by Chahota, Ogawa (34) 13 birds from order Passeriformes were examined and C. psittaci DNA could be demonstrated in one clinically normal Java sparrow. The C. psittaci was, however, typed as genotype D, differing from findings of this study in which the C. psittaci were typed as genotype A. A lower prevalence of C. psittaci is also reported by some other investigators. For instance, Dickx et al. (35) report only 1 of 61 urban pigeons (1.6 %) being PCR-positive for C. psittaci.
The C. psittaci genotypes detected in this study are consistent with the majority of reports in which the predominance genotypes were the genotype B in pigeons (17, 18, 22, 24, 25, 36, 37) and the genotype A in passerines (18, 23).
Other genotypes are also detected in pigeons and passerines. In this regard, a mixed infection of genotypes A, B, and E/B in one pigeon (30), genotype E in a feral pigeon in Switzerland (38), genotype A in three pigeons in Germany (39) and genotypes B, C and D in-home and feral pigeons by Dickx et al. (35), have been reported. Genotype B has also been isolated from canaries (a genus in the Passeriformes order) (23). Geens et al. and Vanrompay et al. also found genotype B to be particularly associated with the pigeon host (17, 25, 40). However, this genotype has been recovered from many bird species, including turkeys, parakeets, and ducks (23).
We were unable to determine the genotype of human PCR-positive samples due to the small amount of DNA and failure of genotyping PCR as relatively low C. psittaci loads could not be amplified by genotyping PCR (20, 25). However, the short DNA sequence obtained from amplicons of the diagnostic PCRs identified the C. psittaci as genotype B. The C. psittaci positive individuals, however, had none of the clinical symptoms of psittacosis such as fever, rigors, sweats, headache, painful joints, and mild cough (41), possibly due to low or nonviable C. psittaci. Genotype B usually causes mild respiratory symptoms, but genotypes A and D, often detected in Passeriformes, can cause serious infections in the human (17, 35, 42).
Kalmar et al. (43) detected viable genotype B in 30% of pharyngeal swabs (3 out of 10) from workers of a refuge center who have had frequent close contact with birds. Other studies have also reported a relatively high incidence of C. psittaci in human populations. Of 540 apparently healthy people who dealt with avian species, 69 (12.7 %) were positive for C. psittaci DNA. In a separate study carried out in Belgium, the number of positive samples in pigeon fanciers was 4 out of 32 (12.5 %), with two C. psittaci samples being genotype D. These data emphasize the potential hazard of free-living birds as a source of C. psittaci infection in humans, especially to those in close contact.
Few studies have been conducted for the detection of C. psittaci in Iran. Mahzounieh et al. (10) showed that 25% of fecal samples in asymptomatic parrots were DNA positive; however, the PCR target gene 16s rRNA and the genotype were not determined. C. psittaci infection in pet birds and pigeons in Iran has been recently studied by molecular techniques (7, 22).
To the best of our knowledge, this study is the first to report molecular characterization of C. psittaci in the human and the most popular free-living birds in this area of the country. Our findings suggest that the pigeons and passerines may harbor C. psittaci and pose as a source of infection for susceptible individuals in public places and parks. Besides the zoonotic potential, there is also the risk of infection of domesticated birds, such as pet birds and poultry, which live in closer contact with human beings. However, more comprehensive studies are required to determine the extent of C. psittaci prevalence and to achieve a broader molecular characterization of C. psittaci in birds as well as healthy and hospitalized individuals so that a precise risk assessment can be made.
Acknowledgements
References
-
1.
Kaleta EF, Taday EM. Avian host range of Chlamydophila spp. based on isolation, antigen detection and serology. Avian Pathol. 2003;32(5):435-61. [PubMed ID: 14522700]. https://doi.org/10.1080/03079450310001593613.
-
2.
Page LA. Comparison of “pathotypes” among Chlamydial (Psittacosis) STRAINS RECOVERED FROM DISEASED BIRDS AND MAMMals. Bull Wildlife Dis Assoc. 1967;3(4):166-75. https://doi.org/10.7589/0090-3558-3.4.166.
-
3.
Longbottom D, Coulter LJ. Animal chlamydioses and zoonotic implications. J Comp Pathol. 2003;128(4):217-44. [PubMed ID: 12834606]. https://doi.org/10.1053/jcpa.2002.0629.
-
4.
Smith KA, Bradley KK, Stobierski MG, Tengelsen LA; National Association of State Public Health Veterinarians Psittacosis Compendium Committee. Compendium of measures to control Chlamydophila psittaci (formerly Chlamydia psittaci) infection among humans (psittacosis) and pet birds, 2005. J Am Vet Med Assoc. 2005;226(4):532-9. [PubMed ID: 15742693]. https://doi.org/10.2460/javma.2005.226.532.
-
5.
Ito I, Ishida T, Mishima M, Osawa M, Arita M, Hashimoto T, et al. Familial cases of psittacosis: Possible person-to-person transmission. Intern Med. 2002;41(7):580-3. [PubMed ID: 12132529]. https://doi.org/10.2169/internalmedicine.41.580.
-
6.
Beeckman DS, Vanrompay DC. Zoonotic Chlamydophila psittaci infections from a clinical perspective. Clin Microbiol Infect. 2009;15(1):11-7. [PubMed ID: 19220335]. https://doi.org/10.1111/j.1469-0691.2008.02669.x.
-
7.
Mahzonieh M, Ghasemi Shamsabadi M, Heidari F. Prevalence of chlamydia psittaci in pigeons in Chaharmahal va Bakhtiari and Yazd provinces of Iran, by nested-PCR, 2012. Iran J Med Microbiol. 2013;7(1):1-6.
-
8.
Kuldeep D, Sandip C, Ruchi T, Singh SD. Avian chlamydiosis (psittacosis/ornithosis): Diagnosis, prevention and control, and its zoonotic concerns. Res Opin Anim Vet Sci. 2013;3(6):157-69.
-
9.
Feng Y, Feng YM, Zhang ZH; et al. Prevalence and genotype of Chlamydia psittaci in faecal samples of birds from zoos and pet markets in Kunming, Yunnan, China. J Zhejiang Uni Sci. 2016;57(10):311. https://doi.org/10.1631/jzus.B1500091.
-
10.
Mahzounieh MR, Heidari Khoei H, Niknejad M, Yektaneh F. Molecular detection of Chlamydia psittaci in fecal samples of asymptomatic parrots in Mazandaran, Iran. Pajoohandeh J. 2014;18(6):337-43.
-
11.
Beckmann KM, Borel N, Pocknell AM, Dagleish MP, Sachse K, John SK, et al. Chlamydiosis in British Garden Birds (2005-2011): Retrospective diagnosis and Chlamydia psittaci genotype determination. Ecohealth. 2014;11(4):544-63. [PubMed ID: 24947738]. [PubMed Central ID: PMC4368850]. https://doi.org/10.1007/s10393-014-0951-x.
-
12.
Haag-Wackernagel D, Moch H. Health hazards posed by feral pigeons. J Infect. 2004;48(4):307-13. [PubMed ID: 15066331]. https://doi.org/10.1016/j.jinf.2003.11.001.
-
13.
Petrovay F, Balla E. Two fatal cases of psittacosis caused by Chlamydophila psittaci. J Med Microbiol. 2008;57(Pt 10):1296-8. [PubMed ID: 18809562]. https://doi.org/10.1099/jmm.0.2008/001578-0.
-
14.
McGuigan CC, McIntyre PG, Templeton K. Psittacosis outbreak in Tayside, Scotland, December 2011 to February 2012. Euro Surveill. 2012;17(22). [PubMed ID: 22687915]. https://doi.org/10.2807/ese.17.22.20186-en.
-
15.
Matsui T, Nakashima K, Ohyama T, Kobayashi J, Arima Y, Kishimoto T, et al. An outbreak of psittacosis in a bird park in Japan. Epidemiol Infect. 2008;136(4):492-5. [PubMed ID: 17559693]. [PubMed Central ID: PMC2870830]. https://doi.org/10.1017/S0950268807008783.
-
16.
Ling Y, Chen H, Chen X, Yang X, Yang J, Bavoil PM, et al. Epidemiology of Chlamydia psittaci infection in racing pigeons and pigeon fanciers in Beijing, China. Zoonoses Public Health. 2015;62(5):401-6. [PubMed ID: 25244602]. https://doi.org/10.1111/zph.12161.
-
17.
Geens T, Desplanques A, Van Loock M, Bonner BM, Kaleta EF, Magnino S, et al. Sequencing of the Chlamydophila psittaci ompA gene reveals a new genotype, E/B, and the need for a rapid discriminatory genotyping method. J Clin Microbiol. 2005;43(5):2456-61. [PubMed ID: 15872282]. [PubMed Central ID: PMC1153750]. https://doi.org/10.1128/JCM.43.5.2456-2461.2005.
-
18.
Sachse K, Laroucau K, Vorimore F, Magnino S, Feige J, Muller W, et al. DNA microarray-based genotyping of Chlamydophila psittaci strains from culture and clinical samples. Vet Microbiol. 2009;135(1-2):22-30. [PubMed ID: 18950965]. https://doi.org/10.1016/j.vetmic.2008.09.041.
-
19.
Wolff BJ, Morrison SS, Pesti D, Ganakammal SR, Srinivasamoorthy G, Changayil S, et al. Chlamydia psittaci comparative genomics reveals intraspecies variations in the putative outer membrane and type III secretion system genes. Microbiology. 2015;161(7):1378-91. [PubMed ID: 25887617]. [PubMed Central ID: PMC4635502]. https://doi.org/10.1099/mic.0.000097.
-
20.
Heddema ER, van Hannen EJ, Duim B, Vandenbroucke-Grauls CM, Pannekoek Y. Genotyping of Chlamydophila psittaci in human samples. Emerg Infect Dis. 2006;12(12):1989-90. [PubMed ID: 17326961]. [PubMed Central ID: PMC3291338]. https://doi.org/10.3201/eid1212.051633.
-
21.
Vanrompay D, Harkinezhad T, van de Walle M, Beeckman D, van Droogenbroeck C, Verminnen K, et al. Chlamydophila psittaci transmission from pet birds to humans. Emerg Infect Dis. 2007;13(7):1108-10. [PubMed ID: 18214194]. [PubMed Central ID: PMC2878242]. https://doi.org/10.3201/eid1307.070074.
-
22.
Madani SA, Peighambari SM. PCR-based diagnosis, molecular characterization and detection of atypical strains of avian Chlamydia psittaci in companion and wild birds. Avian Pathol. 2013;42(1):38-44. [PubMed ID: 23391180]. https://doi.org/10.1080/03079457.2012.757288.
-
23.
Vanrompay D, Butaye P, Sayada C, Ducatelle R, Haesebrouck F. Characterization of avian Chlamydia psittaci strains using omp1 restriction mapping and serovar-specific monoclonal antibodies. Res Microbiol. 1997;148(4):327-33. [PubMed ID: 9765811]. https://doi.org/10.1016/S0923-2508(97)81588-4.
-
24.
Magnino S, Haag-Wackernagel D, Geigenfeind I, Helmecke S, Dovc A, Prukner-Radovcic E, et al. Chlamydial infections in feral pigeons in Europe: Review of data and focus on public health implications. Vet Microbiol. 2009;135(1-2):54-67. [PubMed ID: 18977610]. https://doi.org/10.1016/j.vetmic.2008.09.045.
-
25.
Heddema ER, Ter Sluis S, Buys JA, Vandenbroucke-Grauls CM, van Wijnen JH, Visser CE. Prevalence of Chlamydophila psittaci in fecal droppings from feral pigeons in Amsterdam, The Netherlands. Appl Environ Microbiol. 2006;72(6):4423-5. [PubMed ID: 16751560]. [PubMed Central ID: PMC1489607]. https://doi.org/10.1128/AEM.02662-05.
-
26.
Nei M, Kumar S. Molecular evolution and phylogenetics. New York: Oxford University Press; 2000.
-
27.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-9. [PubMed ID: 24132122]. [PubMed Central ID: PMC3840312]. https://doi.org/10.1093/molbev/mst197.
-
28.
Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39(4):783-91. [PubMed ID: 28561359]. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x.
-
29.
Lewis VJ, Thacker WL, Cacciapuoti AF. Detection of Chlamydia psittaci by immunofluorescence. Appl Microbiol. 1972;24(1):8-12. [PubMed ID: 4626909]. [PubMed Central ID: PMC380536].
-
30.
Geigenfeind I, Vanrompay D, Haag-Wackernagel D. Prevalence of Chlamydia psittaci in the feral pigeon population of Basel, Switzerland. J Med Microbiol. 2012;61(Pt 2):261-5. [PubMed ID: 21921110]. https://doi.org/10.1099/jmm.0.034025-0.
-
31.
Olsen B, Persson K, Broholm KA. PCR detection of Chlamydia psittaci in faecal samples from passerine birds in Sweden. Epidemiol Infect. 1998;121(2):481-4. [PubMed ID: 9825803]. [PubMed Central ID: PMC2809549]. https://doi.org/10.1017/s0950268898001320.
-
32.
de Lima VY, Langoni H, da Silva AV, Pezerico SB, de Castro AP, da Silva RC, et al. Chlamydophila psittaci and Toxoplasma gondii infection in pigeons (Columba livia) from Sao Paulo State, Brazil. Vet Parasitol. 2011;175(1-2):9-14. [PubMed ID: 21055877]. https://doi.org/10.1016/j.vetpar.2010.10.006.
-
33.
Salinas J, Caro MR, Cuello F. Antibody prevalence and isolation of Chlamydia psittaci from pigeons (Columba livia). Avian Dis. 1993;37(2):523-7. [PubMed ID: 8363515].
-
34.
Chahota R, Ogawa H, Mitsuhashi Y, Ohya K, Yamaguchi T, Fukushi H. Genetic diversity and epizootiology of Chlamydophila psittaci prevalent among the captive and feral avian species based on VD2 region of ompA gene. Microbiol Immunol. 2006;50(9):663-78. [PubMed ID: 16985288]. https://doi.org/10.1111/j.1348-0421.2006.tb03839.x.
-
35.
Dickx V, Beeckman DSA, Dossche L, Tavernier P, Vanrompay D. Chlamydophila psittaci in homing and feral pigeons and zoonotic transmission. J Med Microbiol. 2010;59(Pt 11):1348-53. [PubMed ID: 20705727]. https://doi.org/10.1099/jmm.0.023499-0.
-
36.
Horvatek Tomić D, Laroucau K, Prukner-Radovcic E. Detection of Chlamydia psittaci genotypes in fecal samples of homing pigeons in Croatia. Veterinarski Arhiv. 2013;83(2):201-9.
-
37.
Gasparini J, Erin N, Bertin C, Jacquin L, Vorimore F, Frantz A, et al. Impact of urban environment and host phenotype on the epidemiology of Chlamydiaceae in feral pigeons (Columba livia). Environ Microbiol. 2011;13(12):3186-93. [PubMed ID: 21933318]. https://doi.org/10.1111/j.1462-2920.2011.02575.x.
-
38.
Zweifel D, Hoop R, Sachse K, Pospischil A, Borel N. Prevalence of Chlamydophila psittaci in wild birds—potential risk for domestic poultry, pet birds, and public health? Eur J Wildlife Res. 2009;55(6):575-81. https://doi.org/10.1007/s10344-009-0275-2.
-
39.
Sachse K, Kuehlewind S, Ruettger A, Schubert E, Rohde G. More than classical Chlamydia psittaci in urban pigeons. Vet Microbiol. 2012;157(3-4):476-80. [PubMed ID: 22296995]. https://doi.org/10.1016/j.vetmic.2012.01.002.
-
40.
Vanrompay D, Andersen AA, Ducatelle R, Haesebrouck F. Serotyping of European isolates of Chlamydia psittaci from poultry and other birds. J Clin Microbiol. 1993;31(1):134-7. [PubMed ID: 8417017]. [PubMed Central ID: PMC262635].
-
41.
Stewardson AJ, Grayson ML. Psittacosis. Infect Dis Clin North Am. 2010;24(1):7-25. [PubMed ID: 20171542]. https://doi.org/10.1016/j.idc.2009.10.003.
-
42.
Harkinezhad T, Geens T, Vanrompay D. Chlamydophila psittaci infections in birds: A review with emphasis on zoonotic consequences. Vet Microbiol. 2009;135(1-2):68-77. [PubMed ID: 19054633]. https://doi.org/10.1016/j.vetmic.2008.09.046.
-
43.
Kalmar ID, Dicxk V, Dossche L, Vanrompay D. Zoonotic infection with Chlamydia psittaci at an avian refuge centre. Vet J. 2014;199(2):300-2. [PubMed ID: 24326117]. https://doi.org/10.1016/j.tvjl.2013.10.034.