1. Background
Cryptococcus neoformans is an encapsulated yeast pathogen with worldwide distribution, and pigeon droppings are considered an important ecological niche of this pathogen yeast (1-4). Patients with impaired immune systems such as HIV patients, corticosteroid users, and organ transplantation have a higher risk for developing cryptococcosis (5, 6). Inhalation of environmental, infectious propagules or direct contact with etiologic agents can cause cryptococcosis in humans (1). About 70 Cryptococcus species have been identified (7); however, the highest incidence of cryptococcosis was attributed to C. neoformans (var. grubii and var. neoformans) and C. gattii (8, 9). Cryptococcus neoformans var. grubii has a worldwide distribution and alone causes about 80% of cryptococcal infections (6). Whereas C. neoformans var. neoformans is more common in Europe, and C. gattii is common in tropical and subtropical regions (5).
The pathogenicity of Cryptococcus species is associated with two factors, host factors and organism virulence factors. Organism virulence factors consist of capsule and melanin production, growth at 37ºC, and secretion of extracellular enzymes (proteinase, phospholipase, esterase, and urease) (4, 10, 11). Virulence factors are an essential part of microbial pathogenesis. Some of these factors, such as extracellular enzyme production, are common among several species (4). On the other hand, some factors (e.g., capsule formation) are specific to one species (12, 13).
Extracellular enzymes cause host tissue damage and provide optimal conditions for microorganism adhesion to tissues (1, 14). Proteases and phospholipases degrade proteins and glycerophospholipids, respectively, as well as help to colonization and tissue invasion (1, 4, 15). Esterase activity has been associated with virulence factors in Cryptococcus (1); however, its role in tissue invasion remains still unclear. Urease is one of the most necessary enzymes for escaping Cryptococcus from the lungs to the blood-brain barrier (1). Furthermore, C. neoformans induces antioxidant activation enzyme (catalase). Catalase converts hydrogen peroxide to water and molecular oxygen and facilitates the growth and survival of Cryptococcus in macrophages (16).
The capsule has a key role in C. neoformans virulence; thus, capsule-free strains have low or without pathogenicity (10, 11, 13). Melanin is a critical pathogenic factor in C. neoformans that is produced by the phenoloxidase enzyme (17, 18). The function of melanin involves the protection of the fungal cell against oxidative degradation, increases the resistance of fungal cells in tissue and environment, immune modulation, and inflammatory response, and probably increased resistance to amphotericin B drug (1, 19, 20).
2. Objectives
Despite the development of molecular methods, very limited studies have been conducted on the diversity of Cryptococcus species in Iran. Most clinical available reports in Iran are as case reports (21-23), and environmental epidemiological studies are limited to formal identification. The present study aimed to isolate and identify Cryptococcus species from pigeon guano based on molecular methods in Ahvaz, Iran and investigate the important virulence factors in the isolated species.
3. Methods
3.1. Fungal Isolates and Growth Conditions
One hundred and forty-one dried pigeon droppings samples and 76 samples of pigeon cages were collected from 10 different areas in Ahvaz, southwest of Iran. Samples were collected from private houses and pet-shops in sterile packets and transferred to the medical mycology laboratory affiliated to Ahvaz Jundishapur University of Medical Sciences. Approximately 5-10 g of each pigeon dropping was transferred to a sterile tube containing 50 ml of sterile distilled water with several antibiotics, containing penicillin 1,200 U (1 g/L) (Pharmco Jabir Hayyan, Iran), gentamicin (20 µg/L) (Pharmco Jabir Hayyan, Iran) and chloramphenicol (1 g/L) (Bio Basic, Canada). The suspension was mixed for 5 minutes by a vortex (Heidolph, Germany) and allowed to sediment for 1 h, then, 0.1 mL of supernatant was streaked on a niger seed agar (NSA) plate (24).
Samples from cages were collected by rubbing the sterile and moist cotton swabs on the cage walls and immediately plated onto the NSA plates. The cultures were incubated at 32°C and monitored for mucoid and dark-brown colonies for up to two weeks. Then, the Indian ink smears of suspected colonies to Cryptococcus were prepared, and the presence of capsules confirmed Cryptococcus species. Brown colonies were streaked on the new NSA to obtain single and pure colonies. Pure colonies were subcultured on Sabouraud dextrose agar (SDA) (Pronadisa, Spain) slant tubes and stored at room temperature (25-29°C) until use. In this study, a sequenced C. neoformans var. grubii isolate (Accession no = KY216187.1) was gifted by prof. Pakshir (Shiraz University of Medical Sciences, Iran) was used as a positive control for all experiments.
3.2. Identification of Cryptococcus Isolates
3.2.1. Classical Identification
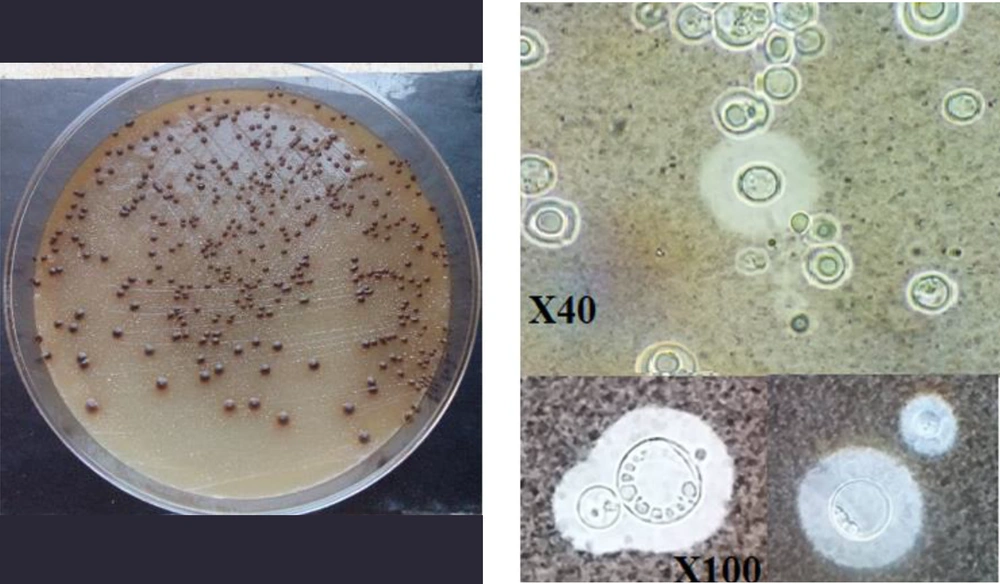
All isolates were initially identified as Cryptococcus spp. by morphological and microcopy features and biochemical tests, including the Indian ink test, thermotolerance at 37°C, melanin synthesis on NSA medium, and urease production (Figure 1) (24, 25).
3.2.2. Molecular Identification
For DNA extraction, each isolate was sub-cultured on SDA and incubated at 28°C for two days. A loopful of fresh yeasts was transferred into a sterile microtube containing 300 ml of lysis buffer and 50 mg glass bead (Sigma-Aldrich, USA) and put at -20°C for 24 h. Microtube contents were homogenized by a SpeedMill PLUS Homogenizer (Analytikjena, Germany) and then were extracted using phenol-chloroform-isoamyl alcohol (Sigma-Aldrich, Germany) (26). The two sets of universal primers, including ITS1 (5'-TCCGTAGGTGAACCTGCGG-3') and ITS4 (5'-TCCTCCGCTTATTGATATGC-3'), were used to amplify the ITS rDNA region (12). The 500 bp bands were observed after the migration of the PCR product by electrophoresis on agarose gel 1.5% (Cinagen, Iran). The PCR products were purified, then sequenced by the Sanger method with primers ITS1 (forward primer), data aligned by MEGA 6 software, and blasted in the NCBI database. The similarity was 100% for 57 (78.1%) isolates whereas 16 (21.9%) isolates had < 99% similarity. All sequenced data were deposited in NCBI GenBank, and accession numbers were obtained.
3.3. Identification of Pathogenic Factors of Cryptococcus Isolates
3.3.1. Capsule Size Determination
A loopful of fresh cultures was inoculated onto diluted Sabouraud dextrose broth (1:10) (BioLife, Italia) (pH = 7.3) and incubated at 37ºC under CO2 condition for 72h (13). Then, 10 µL of a cell suspension was mixed with 10 µL of India ink. A light microscope randomly photographed at least five different fields of each slide. The size of the capsules was scored as large, medium, and small.
3.4. Melanin Production
Thirty microliters of 0.5 McFarland cryptococcal cell suspension were inoculated on NSA and incubated at 30°C for six days (4, 27). Melanin production was evaluated by direct visualization of colony color and scored based on colony color intensity from 1+ to 4+ (4).
3.5. Different Temperature Tolerance
A loopful of each isolate was cultured on both NSA and SDA plates and incubated at 4°C, 30°C, 37°C, 42°C, and 45°C for one month, 48h, 48h, 48h, and 24h respectively. Then, plates were evaluated every day for temperatures 30°C, 37°C, 42°C, and 45°C and 3-4 days for 4°C. Finally, the plates that were previously incubated at 4°C, 42°C, 45ºC, and 30ºC for five days and then were considered for growth again.
3.6. Phospholipase Activity
Secretory phospholipase activity was determined by egg yolk agar medium, according to Price et al. method (28). Egg yolk agar plates were cultivated by isolates and incubated at 30ºC for 15 days in triplicate. Enzyme activity was manifested as a dense zone of precipitation around the colonies, and its level was calculated as precipitation zones (Pz) (27, 29). Pz value is considered by dividing colony diameter by the colony plus precipitation zone. Pz calculated as: Pz value = 1 (negative); Pz value = 0.7-0.99 (weak); Pz value = 0.5-0.69 (mild); and Pz value < 0.5 (strong).
3.7. Proteinase Activity
The proteinase activity of Cryptococcus isolates was assayed based on Aoki et al. methods (30). In brief, 0.1 g KH2PO4 (Merck, Germany), 0.05 g MgSO4, 7 ml H2O (Merck, Germany), 0.01 g yeast extract (Merck, Germany), 1 g glucose (Merck, Germany), and 2 g agar (Mirmedia, Iran) were dissolved in 90 mL distilled water. After sterilization, 10 mL of 0.2% sterilized bovine serum albumin (BSA) (Merck, Germany) was added. The plates were inoculated with isolates and triplicate plates incubated at 30ºC and 37ºC for three weeks. Positive proteinase activity is defined as a clearance zone around the inoculum site. The proteinase activity index (Pz) was calculated as described above.
3.8. Hemolytic Activity
Determination of hemolysin activity was performed by SDA plates containing 3% (W/V) glucose (Merck, Germany) and 7 mL fresh sheep blood (Bahar Afshan, Iran) (V/V%) described by Luo et al. (31). The plates were inoculated with isolates, and triplicate plates were incubated at 30ºC for five days. A translucent halo zone around the colonies indicates the positive hemolytic activity of each isolate. The hemolytic activity index was calculated as described above.
3.9. Esterase Activity
The esterase activity of isolates was evaluated using the described method by Slifkin et al. (32). In brief, the medium contained 10 g peptone (Merck, Germany), 5g NaCl (Mojallali Chemical Laboratories, Iran), 0.1 g CaCl2 (Merck, Germany), 15g agar (Mirmedia, Iran), and 5 mLTween 80 (Merck, Germany) and 1,000 mL distilled water. The esterase medium plates were inoculated with isolates in triplicate and incubated at 30ºC for 15 days. The esterase activity index was calculated as described above.
3.10. Urease Activity
Urease activity of isolates was determined by the urea agar base (Merck, Germany) supplemented by 50 ml/L urea 40%. The medium was inoculated with a 0.5 McFarland standard suspension of each isolate and incubated at 37ºC for one week. The coloration of urea agar was graded from 1+ to 4+, which indicated the intensity of urea hydrolysis (12).
3.11. Catalase Activity
The catalase activity of isolates was detected using an overnight culture. A loopful of a small colony was suspended in an H2O2 drop on a clean microscope slide and monitored to generate Oxygen bubbles.
3.12. Gelatin Hydrolysis
The production of gelatinase was assayed by gelatin agar medium contained 8 g gelatin and 23 g nutrient agar (BioMerieux, France) in 1,000 mL of distilled water. The gelatin agar plates were inoculated with isolates and incubated at 30ºC for four weeks, according to Kanemitsu et al. (33). The presence of a hyaline halo around the inoculated area indicates gelatinase activity.
4. Results
Of 217 samples from pigeon droppings (141 samples) and pigeon cages (76 samples), only 43 (30.5%) pigeon dropping samples were positive for Cryptococcus. Moreover, all cages samples were negative for Cryptococcus. In the present study, 73 Cryptococcus strains were isolated from samples, and based on the sequencing of the ITS rDNA regions, all of the 73 isolates were detected as C. neoformans var. grubii (44 accession numbers: from LC535977 to LC536020; 22 accession numbers: from LC537132 to LC537153; 7 accession numbers: from LC545841 to LC545847). It was found that 30 (41.1%) isolates produced large capsules as well as medium and small capsules, followed by 72 (98.6%) medium capsule size and 66 (90.4%) small capsule size.
All C. neoformans isolates had robust growth at 30ºC and 37ºC, whereas no growth was seen at 45ºC. Moreover, 52 (69.9%) of the isolates could grow at 4ºC, including moderate grow 36 (49.3%), and weak grow 15 (20.6%). When cultures were transferred from 4ºC to 30ºC conditions, most isolates (86.3%) had well growth, followed by moderate growth (12.3%), and weak growth (1.4%). Furthermore, the ability to grow at 42ºC was only found at 14 (19.2%) isolates. Although the melanin intensity and urease activity varied among isolates, 100% of isolates were positive for melanin pigmentation and urease activity (Table 1).
| Activity | Melanin production, No. (%) | Urease activity, No. (%) |
|---|---|---|
| Very high (4+) | 6 (8.2) | 29 (39.7) |
| High (3+) | 24 (32.9) | 26 (35.6) |
| Medium (2+) | 24 (32.9) | 13 (17.8) |
| Low (1+) | 19 (26) | 5 (6.8) |
| Total | 73 (100) | 73 (100) |
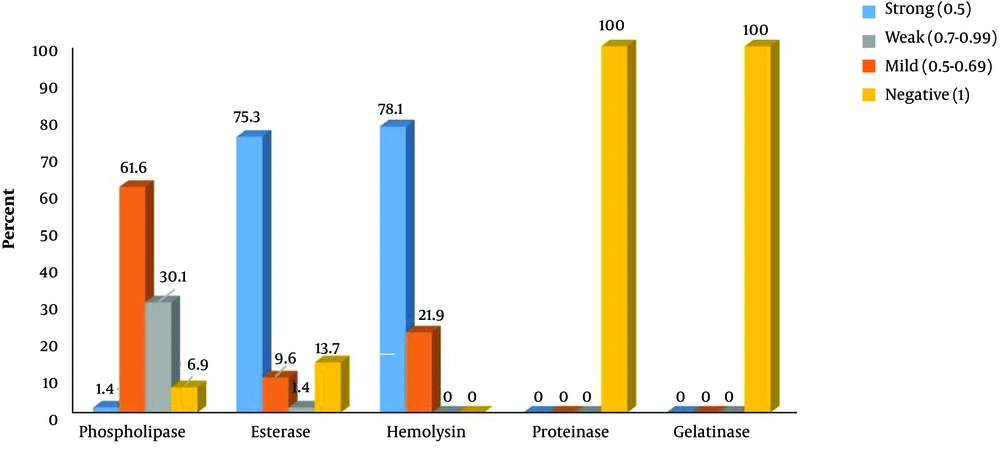
Interestingly, 100%, 93.1%, and 86.3% of isolates produced hemolysin, phospholipase, and esterase extracellular enzymes with different pz index, respectively. Also, all isolates were capable of utilizing hydrogen peroxide. However, all isolates lack extracellular proteinase and gelatinase activity (Figure 2).
5. Discussion
Cryptococcus neoformans has a worldwide distribution, and it is also a common cause of cryptococcosis among patients suffering from AIDS (2, 5, 13, 34). However, few studies have demonstrated the epidemiology of Cryptococcus species and their pathogenic factors in Iran. Moreover, some rare species of Cryptococcus have been isolated in some clinical forms (35). Although some clinical cases of cryptococcosis and a few environmental studies were reported in Iran, C. neoformans var. grubii was only detected by Badali et al., and Pakshir et al., using molecular methods (21-24, 36-38). Cryptococcosis was reported from different parts of the world, including India (39), Italy (40), Brazil (41), and China (7, 42). Besides, C. neoformans var. grubii was the most common variety in clinical and environmental samples (7, 39-43).
The present study is the first epidemiological and molecular identification of Cryptococcus species isolated from pigeon droppings in Ahvaz, southeast Iran. In the present study, 43 (30.5%) pigeon dropping samples were positive for Cryptococcus, and 73 isolates of C. neoformans var. grubii were isolated from samples. Afshari et al. have shown a lower occurrence (5%) of C. neoformans in pigeon excreta in Mazandaran, northern Iran, with the most common species C. neoformans var. grubii (90%) followed by C. neoformans var. neoformans (10%) (5). Although Kamari et al. isolated 12 Cryptococcus species from pigeon nest and Eucalyptus tree samples in Isfahan, only 1 (8.3%) C. neoformans var. grubii was detected (3). Similar to the current study, the frequency of positive cultures for C. neoformans from pigeon droppings was 34%, based on Zarrin et al. report from Ahvaz (44), and they were only identified based on morphological tests. The main source of C. neoformans is pigeon droppings; however, it was also recovered from several trees, including Eucalyptus camaldulensis, Ceratonia siliqua, Olea europaea, and Pinus spp. (45).
Extracellular enzymes have important key roles in the pathogenesis of C. neoformans. These enzymes decompose living host tissues and help invade organs (1, 29). However, the pattern of enzyme secretion varies in different species/strains with different sources (12, 29). Pini et al. believe that there is a significant difference in phospholipase activity between clinical and environmental isolates of C. neoformans (4). In contrast, differences were not found between clinical and environmental isolates of C. neoformans among the evaluated extracellular enzymes in virulence factors in the Andrade-Silva et al. study (41). None of our isolates could secrete proteinase or gelatinase enzymes, while all isolates exhibited a variable rate of hemolysin, catalase, and urease activities. Moreover, the majority of isolates were positive for the presence of phospholipase (93.1%) and esterase (86.3%). Pedroso et al. evaluated the production of phospholipase and urease in C. neoformans var. grubii and found that 100% and 90% of isolates had phospholipase and urease activities, respectively (12). However, they reported that most isolates (54.5%) had proteinase activity.
The ability to form a polysaccharide capsule in C. neoformans is one of the most important virulence factors. In the host body, the capsule prevents phagocytes and protects against oxidative bursts (1, 46). Although higher levels of CO2 stimulate increasing capsule size in C. neoformans strains, CO2 levels have not the same effect on all strains of C. neoformans (13). In the present study, different capsule sizes were observed during incubation at CO2 and 37°C for three days. Large capsules were produced in 41.1% of isolates. Although all of the isolates (100%) were positive for melanin production on G. abyssinica medium, it was found that they differed in the intensity of melanin formation. The melanization in Cryptococcus species is mainly associated with adaptation to adverse environmental conditions and resistance to oxidative damage and antimicrobial compound in tissue cells (19, 47). As a result, melanization plays a vital role in the pathogenicity of Cryptococcus species (17, 18). Pini et al. have shown that 2.4% and 3.1% of clinical strains and environmental strains were negative for melanization, respectively (4).
Although Cryptococcus species can grow in routine media at 25 to 37°C, the ability to grow at 37°C is very important for its pathogenicity (12, 48). In the present study, the isolates' growth was well and similar at temperatures of 30°C and 37°C. Moreover, all incubated plates at 4°C for one month began to grow after incubation at 30°C. Only 19.2% of isolates retained their ability to grow at 42°C and the rest of them (80.8%) were unable to grow at 42°C even after transfer to 30ºC. However, all incubated isolates at 45ºC for 24 h were unable to grow after transfer to 30ºC. Cryptococcus neoformans can produce different heat shock protein (hsp) types (examples, hsp 60, 70, and 80) that protect the organism in mammalian hosts (48).
5.1. Conclusions
Although two methods were used for recovery of Cryptococcus, only Cryptococcus was isolated from pigeon guano, and swabs from the cage walls were negative. Cryptococcus neoformans var. grubii was the only species from pigeon droppings from Ahvaz with more pathogenic factors. Owing to the high pathogenicity of the isolates, the frequency of the disease is expected to be higher.