Abstract
Background:
To date, more than 300,000 distinct peptides have been identified in scorpion venom. However, only a limited number of these peptides have been described.Objectives:
We characterized some putative peptides from the venom gland cDNA library of the Iranian yellow scorpion Hemiscorpius lepturus.Methods:
Total RNA was extracted from yellow Iranian scorpion glands. Single-stranded cDNA (sscDNA) and double-stranded cDNA (dscDNA) were synthesized by polymerase chain reaction (PCR). A cDNA library was achieved by inserting dscDNA into a special vector and subsequently transformed to chemically competent Escherichia coli as a host. The library was screened by culturing the liquid library on Lysogeny broth (LB)-agar plates. Analysis of positive clones was performed by plasmid extraction and the sequencing of the inserts. Finally, all cDNA sequences were analyzed and characterized by bioinformatics software.Results:
One hundred colonies were randomly analyzed. Eighty-nine cDNA sequences had acceptable quality for bioinformatics analysis. Five sequences were selected for further analysis. The peptides related to these sequences were divided into two groups, non-disulfide bridge peptides (NDBP) and disulfide bridge peptides (DBP), the application of which in health and medical issues has been suggested.Conclusions:
The data obtained in this study may be an important resource for further in vivo and in vitro functional assays to identify valuable therapeutic peptides.Keywords
cDNA Library Iranian Scorpion Hemiscorpius lepturus Toxin Pharmaceutical Properties
1. Background
Scorpions, ancient arthropods belonging to the Arachnids class, have lived for over 400 million years (1). More than 1500 species of scorpions are distributed in the vast territory of the world from Asia to Africa, Australia, and America (2). The venom of scorpions is the result of evolution over millions of years and does not exceed 5% of the scorpion’s dry weight. It is a cocktail of peptides, proteins, lipids, nucleotides, mocupolypeptides, biological amines, and other unknown components which could cause immunological and toxicological responses (3). Ion channel (Na, K, Ca, and Cl) blockers are the most critical peptides of scorpion venom (4). During the past two decades, various pharmaceutical properties, including antimicrobial (5, 6), antitumor/anticancer (7), antiepileptic (8), antihypertensive, anti-inflammatory, and anti-osteoporotic effects, as well as treating cardiovascular and autoimmune diseases have been revealed for some peptides and proteins originating from the scorpion venom (9, 10).
Hemiscorpius lepturus, which belongs to the Liochelidae family, is distributed in Iran (specially Khuzestan province), Pakistan, Iraq (Mandeli in the east of Baghdad), and Yemen (11). Hemiscorpius lepturus is the most dangerous scorpion in Iran, which causes 95% of deaths due to scorpion envenomation (12). The venom of this scorpion contains different proteins with molecular weights ranging from 3.5 kD to 260 kD (13).
Studies on H. lepturus sting both in humans and animals showed that this venom includes hemolytic toxins, phospholipases, lactate dehydrogenase (14), hyaluronidases, gelatinase, caseinase (13), neurotoxins, phosphodiesterase, nephrotoxins, serotonin, histamine, glycosaminoglycans, tryptophan, and cytokines release inducers (15). Hemiscorpius lepturus sting does not cause immediate pain at the site of the bite because it damages the presynaptic sensory nerves and inhibits the release of neurotransmitters. However, it provokes delayed cutaneous manifestations, such as swelling, necrosis, macular erythema, bullae, purpuric changes, and ulcers which develop over time, followed by systemic signs (16, 17).
There are few investigations on the components of H. lepturus venom. Borchani, in 2011 isolated a powerful hemolytic and dermonecrotic agent from H. lepturus venom with a molecular weight of 33 kD, named Heminecrolysin (18). In 2007, Shahbazzadeh introduced hemicalcin with some effects on ryanodine-sensitive Ca2+ channels (19). The other toxin of H. lepturus is a potassium channel blocker named hemitoxin that inhibits Kv1.3, Kv1.2, and Kv1.3 in Xenopus laevis oocytes (20).
Over the past two decades, improving proteomic and transcriptomic approaches has provided facilities to identify various components of the venom of animals. Construction of a cDNA library from the venom gland of a scorpion and cloning it in bacteria is a strong instrument to identify the coding sequences of venom toxins, such as neurotoxins and enzymes.
2. Objectives
In this project, the cDNA sequences of the peptides and proteins of the H. lepturus venom gland were synthesized and cloned in a proper vector. Finally, five coding regions related to novel peptides were selected for further evaluation using molecular and bioinformatics analyses. This research provides the framework for detecting the sequence of peptide transcripts and the expression of H. lepturus venom gland peptides, which would provide a useful source of novel pharmacological data for studying ion channels and antimicrobial peptides (AMPs). Moreover, these data help understand the physiological effects of toxins in H. lepturus venom at the molecular levels.
3. Methods
Hemiscorpius lepturus scorpions were captured from the desert around Khuzestan province, southwest of Iran. They were taken to the laboratory of the Toxicology Research Center of Ahvaz Jundishapur University of Medical Sciences (AJUMS) to confirm the species. Animal procedures followed the guidelines for animal care prepared by the Committee on Care and Use of Laboratory Animal Resources, National Institutes of Health, USA (NIH publication 86-23 revised 1985), and were approved by the Animal Ethics Committee of AJUMS by the ethical approval code IR.AJUMS.REC.1396.916.
The milking of toxic glands was carried out by electroshock. Venom glands were cut four days after milking in order to stimulate the synthesis of mRNAs. Venom glands were kept in RNALaterTM (Qiagen). The total RNA was extracted using the protocol described by RNeasy Plus Mini Kit (Qiagen). The full-length cDNA library was constructed by the In-Fusion SMARTer Directional cDNA Library Constriction (Clontech) kit. Double-stranded (ds) cDNA was cloned into pSMARTE2IFD (2716bp) plasmid. Recombinant plasmids were transformed into chemically competent Escherichia coli DH5α cells as previously described. DH5-Alpha cells are engineered E. coli cells to maximize transformation efficiency. Transformant cells were cultured on a solid culture plate. Next, the plasmids of white colonies were extracted by QIAprep Spin Miniprep Kit (Qiagen).
Ten plasmids were randomly selected for sequencing and were sent to Microgen Company.
Obtained sequences were searched against National Center for Biotechnology Information (NCBI) GenBank to find similar sequences. Amino acid sequences were obtained using Open Reading Frame (ORF) Finder program (http://www.ncbi.nlm.nih.gov/projects/gorf/). Protein and nucleotide blasts were performed to identify the putative conserved domains (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Amino acid alignments and the phylogenic trees of predicted protein sequences were obtained from http://www.uniprot.org. The theoretical average molecular mass for putative peptides was calculated by http://web.expasy.org/cgibin/compute_pi/pi_tool. Signal peptide and cleavage sites were predicted by http://www.csbio.sjtu.edu.cn/bioinf/Signal-3L and http://www.cbs.dtu.dk/services/SignalP. Other analyses, such as predicting the tertiary structure of putative proteins and the position of disulfide bridges, were completed by http://swissmodel.expasy.org and http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index, respectively. Finally, the 3-D structures of proteins were displayed by Visual Molecular Dynamics version 1.9.3.
4. Results and Discussion
More than 200 independent white colonies grew on the plate. Colonies were screened, and plasmids were extracted and sent for nucleotide sequencing. From all received sequences, 89 had acceptable ORFs. Sixteen ORFs showed no similarity with known peptides. In contrast, their cDNA sequences in the nucleotide blast showed similarity to the nucleotide sequences of proteins derived from other scorpion species and arthropods. We observed that 52 putative peptide sequences were similar to some peptides, including toxins, AMPs, and cell process proteins originating from scorpions or other arthropods, such as spiders and horseshoes.
The lengths of deduced protein sequences were 60 - 229 amino acids. Furthermore, 59 expressed sequence tags (ESTs) were categorized functionally into five categories of toxins, venom proteins, AMPs, cell proteins, and ESTs with no founded ORF. Herein, six sequences were selected for further analysis. These peptides were divided into two groups, non-disulfide bridge peptides (NDBPs) and disulfide bridge peptides (DBPs). They were named HLx, “H” for Hemiscorpius, “L” for lepturus, and” x” for the number of clone types in the cDNA library.
4.1. Disulfide Bridge Peptides
4.1.1. HL1: A Single Von Willebrand Factor C-domain Proteins (SVWC)
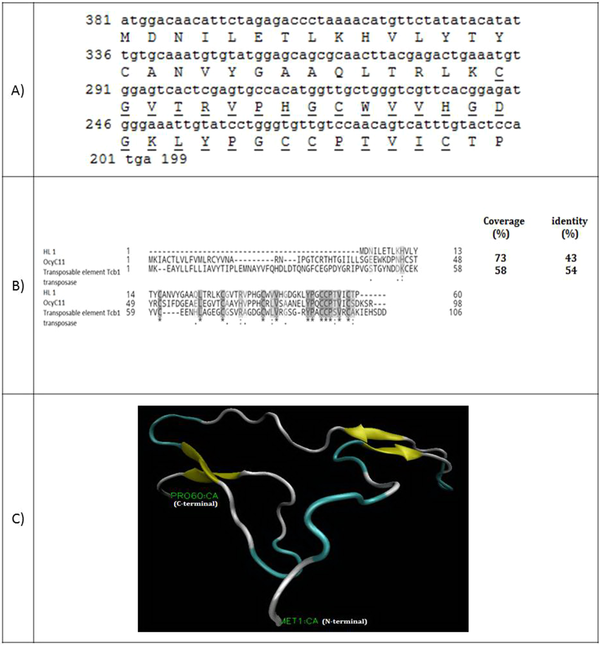
Figure 1A shows the full-length cDNA and amino acid sequences of HL1. The coding sequence of HL1 has 182 nucleotides encoding a 60-amino acid peptide with a molecular weight of 6580.76 Da. Amino acids alignment of HL1 and two similar peptides (OcyC11 and transposable element Tcb1 transposase) are shown in Figure 1B. HL1 is similar to OcyC11 with 73% identity and to transposable element Tcb1 transposase with 54% identity. OcyC11 is a venom toxin isolated from the cDNA library of scorpion Opisthacanthus cayaporum of Liochelidae family, while transposable element Tcb1 transposase originates from spider Stegodyphus mimosarum of Eresoidea family. No function was reported for the OcyC11 venom peptide until now (21). Residues 30 - 58 of HL1 forms an SVWC (Single domain von Willebrand factor type C) domain. This is a conserved domain present in lower eukaryotes and dominantly in arthropods. SWVC proteins are responsible for environmental challenges, such as bacterial infection and nutritional status evoking anti-viral immunity (22). The 3-D structure of HL1 was determined by the Phyre2 server (Figure 1C). This structure consists of three β-strands linked to each other by three disulfide bridges between cysteine residues (58 and 39), (53 and 30), and (52 and 16).
(A) Coding sequence and amino acid sequence of HL1. The amino acids of SWC peptide are underlined; (B) Alignment of venom toxin OcyC11 from Opisthacanthus cayaporum and transposable element Tcb1 transposase with HL1. Coverage and identity percent have been mentioned; (C) The molecular structure of HL1 peptide using the modeling software
4.1.2. HL4: A Potassium Channel Toxin (KTx)
The full-length cDNA of HL4 has 200 nucleotides and encodes a 66 amino-acid peptide with a molecular weight of 7361.39 Da (Figure 2A). A 20 amino-acid signal peptide was identified. Figure 2B shows the amino acids alignment of HL4 and four known similar scorpion peptides, including kappa-KTx 2.6 and 2.5 from scorpion Opisthacanthus cayaporum (Liochelidae family) and kappa-KTx 2.7 and 2.8 from Heterometrus petersii (Scorponidae family). Scorpion toxins that affect potassium channels (KTxs) are divided into four groups: α-, β-, γ-, and κ-KTxs. Kappa type of KTxs is found in Liochelidae and Scorpionidae families (23).
(A) Coding sequence of HL4 and its predicted amino acids. The amino acids of the signal peptide are bold and amino acids of transmembrane helixes are underlined; (B) Alignment of HL4 and four potassium channel blockers of different scorpions. Coverage and identity percent have been mentioned; (C) Phylogenic tree of HL4; (D) The structure of HL4
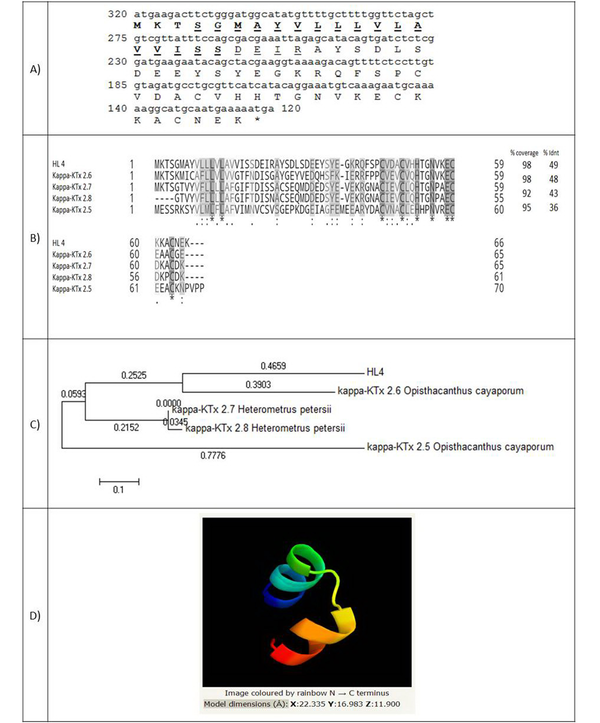
Figure 2C indicates the phylogenic tree of HL4, which was drawn based on the similarity of HL4 with four KTxs from Liochelidae and Scorpionidae families (E-value < 1) by MEGA 5.10 software. The lengths of horizontal lines are estimated. HL4 is near to kappa-KTx 2.6 of Opisthacanthus cayaporum, which, together with Hemiscorpius, are from the Liochelidae family.
The 3-D structure of HL4 was identified by Phyre2 software based on 17 templates (Figure 2D). Different folding types of the 3-D structure have been reported for κ-KTxs up to now: αα, βββ, αββ, ββ, ββα, and βαββ (24, 25). HL4 model is αα type by two parallel alpha helixes, linked by two disulfide bridges through cysteine residues (45 and 63) and (49 and 59) stabilizing the protein structure. The αα folding type is specific for the kappa type of KTxs (23). Although KTxs are K+ channels-selective, κ-KTxs with αα folding can also inhibit voltage-gated Na+ channels (26).
4.2. Non-disulfide Bridge Containing Proteins
4.2.1. HL2: A Pentraxin-like Protein
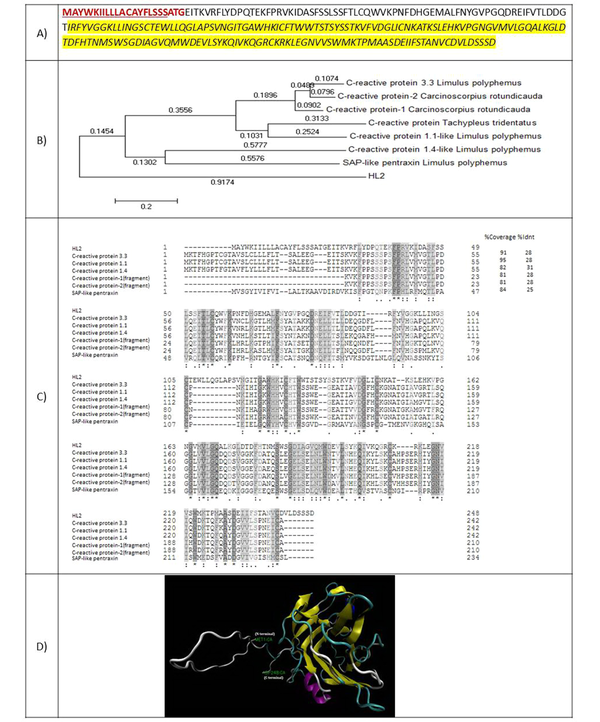
HL2 is a protein with 248 amino acids and a molecular weight of 27576.66 Da. The amino acid sequence of this protein showed 28% identity with the C-reactive protein of Limulus polyphemus horseshoe (Figure 3A). Residues 56 - 235 belong to the Laminin G domain superfamily, and residues 56 - 224 were aligned with the pentraxin family. A 23-amino acid signal peptide was identified for HL2. Figure 3B presents the phylogenic tree of HL2 in comparison with some other pentraxins. Figure 3C shows the amino acid alignment of HL2 with known similar proteins. Figure 3D demonstrates three 3-D structures predicted for HL2 using the model of a serum amyloid protein-like protein belonging to the pentraxin family.
(A) Amino acid sequence of HL2. The amino acids of the signal peptide are shown in red and bold. The amino acids of transmembrane helices are underlined. Amino acids of C-reactive protein from the pentraxin family are highlighted by yellow; (B) Phylogenic tree of HL2; (C) amino acid alignment of HL2 with known similar proteins; (D) 3-D structure of HL2.
4.2.2. HL3: A SIP1-like Protein
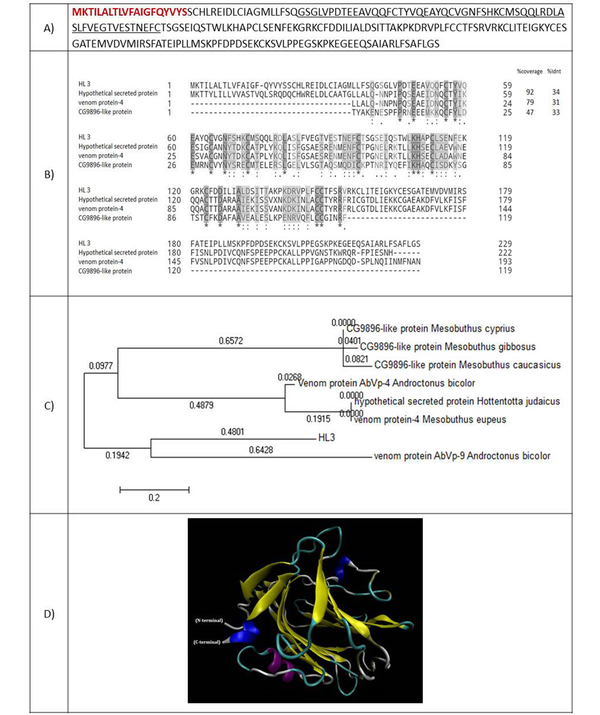
The full-length cDNA of HL3 encodes a 229 amino acid protein. The molecular weight of 25414.19 Da was estimated for this protein (Figure 4A). The amino acids sequence of HL3 was aligned to three similar proteins, including a hypothetical secreted protein from scorpion Hottentotta judaicus with an identity of 34%, a venom protein of scorpion Mesobuthus eupeus with an identity of 31%, and the CG9896-like protein of Mesubutus eupeus with an identity of 33% (Figure 4B). Residues 40 - 96 of HL3 matched to survival motor neuron interacting protein 1 (SIP1) family. A 20-amino acid signal peptide was predicted for HL3. Figure 4C shows the phylogenic tree of HL3 and nine similar putative scorpions' proteins. Figure 4D presents the predicted 3-D structure of HL3 using ten templates. The first and third models are the repulsive guidance molecule (RGM) domain family member B, and the second is a model of C-reactive RGM A.
(A) Amino acid sequence of HL3. The amino acids of the signal peptide are shown in red and bold. The amino acids of SIP1 protein are underlined; (B) Alignment of HL3 with similar peptides. Coverage and identity percent have been mentioned; (C) Phylogenic tree of HL3; (D) 3-D structure of HL3.
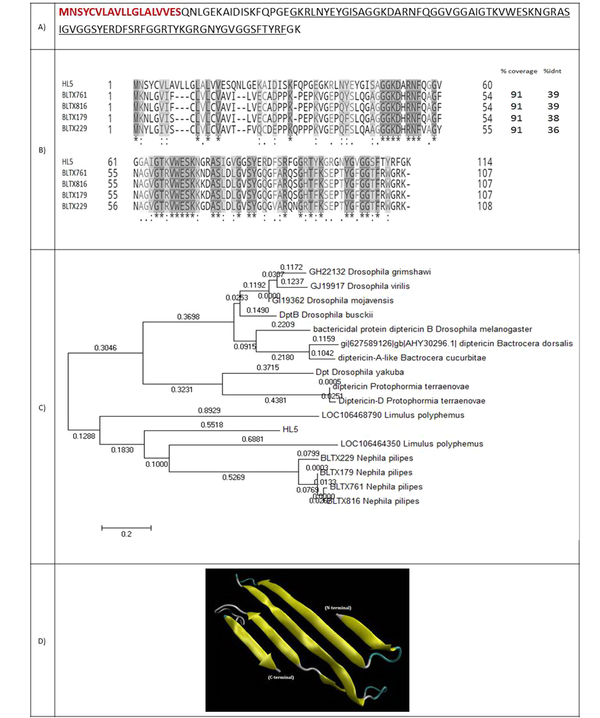
4.2.3. HL5: An Antimicrobial Protein
HL5 is a 114-amino acid protein with a molecular weight of 12129.62 Da (Figure 5A). A 19-amino acid signal peptide was identified for this protein. HL5 showed 39% identity with attacin, an antimicrobial protein from the spider Nephila pilipes. Residues 37-112 aligned to an attacin-c antibiotic. This family includes attacin, sarcotoxin, and diptericin. All the members of this family are insect antibacterial proteins induced by the fat body. They are subsequently released into secreted hemolymph, where they act synergistically to kill the invading microorganism (Figure 5B). Figure 5C shows the phylogenic tree of HL5 and 20 attacin proteins with E-value < 1. Figure 5D indicates the predicted model of the 3-D structure of HL5.
(A) Coding sequence and amino acid sequence of HL5. The amino acids of the signal peptide are shown in red and bold. Amino acids of attacin are underlined; (B) Alignment of HL5 and four spider peptides from the attacin family. Coverage and identity percent have been mentioned; (C) Phylogenic tree of HL5; (D) 3-D structure of HL5.
5. Conclusions
With progress in the field of biotechnology and the use of new transcriptomic methods and proteomic analysis, as well as gene cloning, the number of identified scorpion venom components is rising (27). Here, we identified and described the characteristics of some new proteins, including toxic or non-toxic, expressed in the venom gland of Iranian H. lepturus. Considering the importance of identifying scorpion components in the production of anti-venom medications, the results of the present study can be a guiding light for further studies in the aforementioned field.
Acknowledgements
References
-
1.
Polis GA. The biology of scorpions. Stanford University Press; 1990.
-
2.
Dehghani R, Valaei N. Scorpion bite in Iran: Review of the literature. Feyz J Kashan Univ Med Sci. 2012;1(3).
-
3.
Zhijian C, Feng L, Yingliang W, Xin M, Wenxin L. Genetic mechanisms of scorpion venom peptide diversification. Toxicon. 2006;47(3):348-55. [PubMed ID: 16387337]. https://doi.org/10.1016/j.toxicon.2005.11.013.
-
4.
Baradaran M, Jalali A, Naderi Soorki M, Galehdari H. A Novel Defensin-Like Peptide Associated with Two Other New Cationic Antimicrobial Peptides in Transcriptome of the Iranian Scorpion Venom. Iran Biomed J. 2017;21(3):190-6. [PubMed ID: 27794585]. [PubMed Central ID: PMC5392222]. https://doi.org/10.18869/acadpub.ibj.21.3.190.
-
5.
Baradaran M, Jalali A, Jolodar A, Ghasemian S. A new caerin-like antibacterial peptide from the venom gland of the Iranian scorpion Mesobuthus eupeus: cDNA amplification and sequence analysis. Afr J Biotech. 2012;11(44):10176-81. https://doi.org/10.5897/ajb11.3373.
-
6.
Baradaran M, Jolodar A, Jalali A, Navidpour S, Kafilzadeh F. Sequence analysis of lysozyme C from the scorpion mesobuthus eupeus venom glands using semi-nested rt-PCR. Iran Red Crescent Med J. 2011;13(10):719-25. [PubMed ID: 22737410]. [PubMed Central ID: PMC3371883].
-
7.
Ma R, Mahadevappa R, Kwok HF. Venom-based peptide therapy: insights into anti-cancer mechanism. Oncotarget. 2017;8(59):100908-30. [PubMed ID: 29246030]. [PubMed Central ID: PMC5725072]. https://doi.org/10.18632/oncotarget.21740.
-
8.
Chen Q, Yang P, Lin Q, Pei J, Jia Y, Zhong Z, et al. Effects of scorpion venom heat-resistant peptide on the hippocampal neurons of kainic acid-induced epileptic rats. Braz J Med Biol Res. 2021;54(5). e10717. [PubMed ID: 33825777]. [PubMed Central ID: PMC8021226]. https://doi.org/10.1590/1414-431X202010717.
-
9.
Suhas R. Structure, function and mechanistic aspects of scorpion venom peptides - A boon for the development of novel therapeutics. Eur J Med Chem Rep. 2022;6:100068. https://doi.org/10.1016/j.ejmcr.2022.100068.
-
10.
Ortiz E, Gurrola GB, Schwartz EF, Possani LD. Scorpion venom components as potential candidates for drug development. Toxicon. 2015;93:125-35. [PubMed ID: 25432067]. [PubMed Central ID: PMC7130864]. https://doi.org/10.1016/j.toxicon.2014.11.233.
-
11.
Lourenço WR. The scorpion families and their geographical distribution. J Venom Anim Toxins. 2001;7(1):3-23. https://doi.org/10.1590/s0104-79302001000100002.
-
12.
Alavi SM, Alavi L. Epidemiology of animal bites and stings in Khuzestan, Iran, 1997-2006. J Infect Public Health. 2008;1(1):51-5. [PubMed ID: 20701846]. https://doi.org/10.1016/j.jiph.2008.08.004.
-
13.
Seyedian R, Pipelzadeh MH, Jalali A, Kim E, Lee H, Kang C, et al. Enzymatic analysis of Hemiscorpius lepturus scorpion venom using zymography and venom-specific antivenin. Toxicon. 2010;56(4):521-5. [PubMed ID: 20493200]. https://doi.org/10.1016/j.toxicon.2010.05.008.
-
14.
Khodadadi A, Pipelzadeh MH, Vazirianzadeh B, Pipelzadeh M, Sharifat M. An in vitro comparative study upon the toxic properties of the venoms from Hemiscorpius lepturus, Androctonus crassicauda and Mesobuthus eupeus scorpions. Toxicon. 2012;60(3):385-90. [PubMed ID: 22569320]. https://doi.org/10.1016/j.toxicon.2012.04.348.
-
15.
Karalliedde L. Animal toxins. Br J Anaesth. 1995;74(3):319-27. [PubMed ID: 7718380]. https://doi.org/10.1093/bja/74.3.319.
-
16.
Pipelzadeh MH, Jalali A, Taraz M, Pourabbas R, Zaremirakabadi A. An epidemiological and a clinical study on scorpionism by the Iranian scorpion Hemiscorpius lepturus. Toxicon. 2007;50(7):984-92. [PubMed ID: 17854855]. https://doi.org/10.1016/j.toxicon.2007.07.018.
-
17.
Radmanesh M. Cutaneous manifestations of the Hemiscorpius lepturus sting: a clinical study. Int J Dermatol. 1998;37(7):500-7. [PubMed ID: 9679690]. https://doi.org/10.1046/j.1365-4362.1998.00386.x.
-
18.
Borchani L, Sassi A, Shahbazzadeh D, Strub JM, Tounsi-Guetteti H, Boubaker MS, et al. Heminecrolysin, the first hemolytic dermonecrotic toxin purified from scorpion venom. Toxicon. 2011;58(1):130-9. [PubMed ID: 21658401]. https://doi.org/10.1016/j.toxicon.2011.05.016.
-
19.
Shahbazzadeh D, Srairi-Abid N, Feng W, Ram N, Borchani L, Ronjat M, et al. Hemicalcin, a new toxin from the Iranian scorpion Hemiscorpius lepturus which is active on ryanodine-sensitive Ca2+ channels. Biochem J. 2007;404(1):89-96. [PubMed ID: 17291197]. [PubMed Central ID: PMC1868827]. https://doi.org/10.1042/BJ20061404.
-
20.
Srairi-Abid N, Shahbazzadeh D, Chatti I, Mlayah-Bellalouna S, Mejdoub H, Borchani L, et al. Hemitoxin, the first potassium channel toxin from the venom of the Iranian scorpion Hemiscorpius lepturus. FEBS J. 2008;275(18):4641-50. [PubMed ID: 18699777]. https://doi.org/10.1111/j.1742-4658.2008.06607.x.
-
21.
Silva EC, Camargos TS, Maranhao AQ, Silva-Pereira I, Silva LP, Possani LD, et al. Cloning and characterization of cDNA sequences encoding for new venom peptides of the Brazilian scorpion Opisthacanthus cayaporum. Toxicon. 2009;54(3):252-61. [PubMed ID: 19379768]. https://doi.org/10.1016/j.toxicon.2009.04.010.
-
22.
Chen YH, Jia XT, Zhao L, Li CZ, Zhang S, Chen YG, et al. Identification and functional characterization of Dicer2 and five single VWC domain proteins of Litopenaeus vannamei. Dev Comp Immunol. 2011;35(6):661-71. [PubMed ID: 21256150]. https://doi.org/10.1016/j.dci.2011.01.010.
-
23.
Camargos TS, Restano-Cassulini R, Possani LD, Peigneur S, Tytgat J, Schwartz CA, et al. The new kappa-KTx 2.5 from the scorpion Opisthacanthus cayaporum. Peptides. 2011;32(7):1509-17. [PubMed ID: 21624408]. https://doi.org/10.1016/j.peptides.2011.05.017.
-
24.
Jouirou B, Mouhat S, Andreotti N, De Waard M, Sabatier JM. Toxin determinants required for interaction with voltage-gated K+ channels. Toxicon. 2004;43(8):909-14. [PubMed ID: 15208024]. https://doi.org/10.1016/j.toxicon.2004.03.024.
-
25.
Mouhat S, Jouirou B, Mosbah A, De Waard M, Sabatier JM. Diversity of folds in animal toxins acting on ion channels. Biochem J. 2004;378(Pt 3):717-26. [PubMed ID: 14674883]. [PubMed Central ID: PMC1224033]. https://doi.org/10.1042/BJ20031860.
-
26.
Barnham KJ, Dyke TR, Kem WR, Norton RS. Structure of neurotoxin B-IV from the marine worm Cerebratulus lacteus: a helical hairpin cross-linked by disulphide bonding. J Mol Biol. 1997;268(5):886-902. [PubMed ID: 9180379]. https://doi.org/10.1006/jmbi.1997.0980.
-
27.
Quintero-Hernandez V, Jimenez-Vargas JM, Gurrola GB, Valdivia HH, Possani LD. Scorpion venom components that affect ion-channels function. Toxicon. 2013;76:328-42. [PubMed ID: 23891887]. [PubMed Central ID: PMC4089097]. https://doi.org/10.1016/j.toxicon.2013.07.012.
