1. Background
Cryptosporidium parvum parasite is intracellular and can cause diarrhea (1). Considering the fact that the parasite slows down a wide range of hosts, it can be thought of as a pathogen shared by humans and livestock (2). Soil, water, and food contaminated with human or animal infectious stools are among the most important factors in the transmission of parasites (3).
The infection is self-limiting in healthy individuals (immunocompetent), yet the prolonged disease course is associated with chronic diarrhea, severe dehydration, vomiting, colic, and severe weight loss in people with a weakened immune system, such as those with AIDS, transplantation recipients, those undergoing corticosteroid therapy, IgA deficiency, malnutrition, and Hodgkin’s patients (4, 5). In most cases, water is the source of infection (6). Cryptosporidium in the World Health Organization’s (WHO) pathogens reference list is considered as one of the indicators for assessing global water quality (7). The spread of this disease has not been limited by geographical boundaries and is widely dispersed across the globe (8, 9). With the advent of the AIDS phenomenon in the 1980s, this single-cell protozoan became increasingly important (10). The prevalence of Cryptosporidium is estimated to be between 1% and 3% in European and North American countries, 5% in Asia, and 10% in Africa (11). In a study from France, the prevalence rate was 21.1% in HIV-positive individuals (11). Molecular tools are used for the epidemiology of cryptosporidiosis, classification, biology, and dedicated host of each species, as well as the study of the genetic diversity between Cryptosporidium. Species identification helps determine the sources of infection and transmission, and important human pathogens and their pathogenicity. In Iran, infection with Cryptosporidium spp. has been reported and molecular genetics was done to differentiate species and genotype discrimination of the oocyst (12). The molecular assays indicated that Cryptosporidium parvum is the most common species found in Iran (84.4%), followed by C. hominis (13.74%) (13). Only one study indicated that C. meleagridis has been found in a child in Mazandaran province, northern Iran (13). In some studies, data show that Cryptosporidium prevalence in children under five years old was significantly higher than children above five years old (8, 14, 15).
2. Objectives
Considering that no study has been carried out in this area so far, the present research was conducted to determine the genetic diversity of Cryptosporidium in children with diarrhea in Zabol, using 18srRNA genes and the PCR-RFLP method.
3. Methods
Human specimens were obtained from children with diarrhea, who had been referred to Amir-Almomenin Hospital, Imam Khomeini Hospital and the Central Laboratory of Zabol. After transferring the specimen to the university’s laboratory with direct methods, a thin smear of the specimens was prepared on a slide, stained using Ziehl–Neelsen stain method. The specimens were later investigated in terms of the presence of Cryptosporidium oocyst and the purified positive samples were kept at freezer - 20°C until DNA extraction, using the smear method.
3.1. DNA Extraction
Specimen suspensions were washed three to six times using PBS before the transferring process to perform freeze-thaw and PCR. Then, the freeze-thaw process was performed three times for 10 minutes in each cycle. DNA extraction was performed using a DNA extraction kit (Yekta Tajhiz), according to the instructions and the DNA concentration extracted by spectrophotometry was measured and kept in the freezer until the PCR.
3.2. PCR
The forward primer pair of GGAAGGGTTGTATTTATTAGATAAAG and reverse primer pair of AAGGAGTAAGGAACAACCTCCA were used to perform PCR for the SSUrRNA gene. Polymerase Chain Reaction was carried out under the following conditions: 35 cycles, initial denaturation at 94°C for five minutes, denaturation at 94°C for 45 seconds, annealing at 55°C for 45 seconds, elongation at 72°C for 60 seconds, and ultimate elongation at 72°C for seven minutes.
3.3. RFLP
Agarose gel electrophoresis method was used to evaluate the PCR results and to ensure the proliferation of the desired fragment. The molecular weight of the intended fragment was determined alongside a DNA marker. The Vsp1 enzyme was used to determine the species and genotypes of Cryptosporidium by SSUrRNA gene. To perform RFLP, the main mixture was reached to a final volume of 31 µL using a 2-μL buffer, 10 µL PCR product, and one unit of the enzyme with distilled water. It was later placed at 37°C in a water bath for two hours. The contents of the product were then electrophoresed on 2% gel. The intended bands were observed along with the DNA marker, using a duct gel device.
4. Results
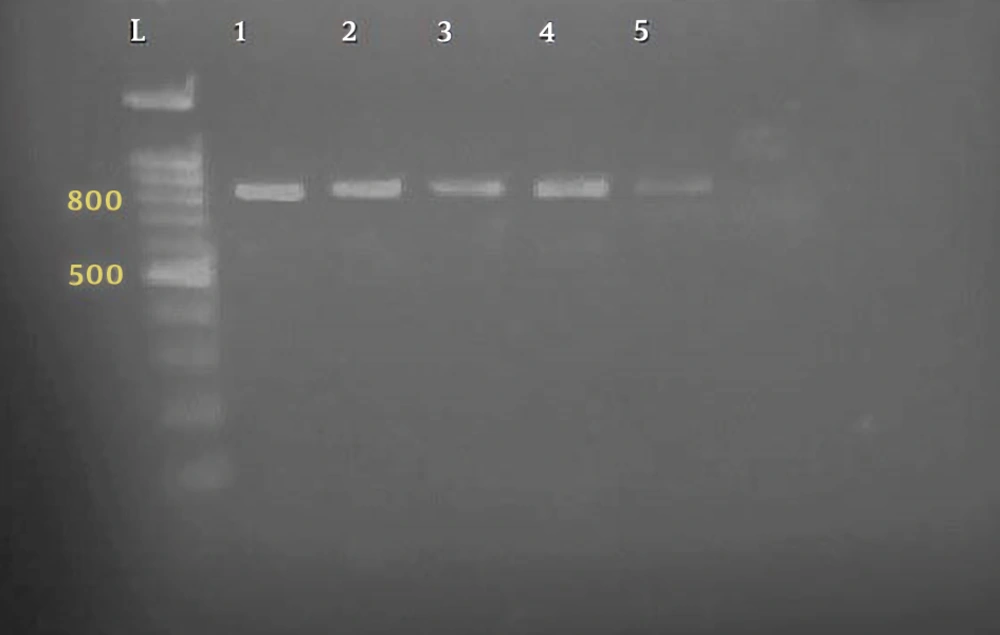
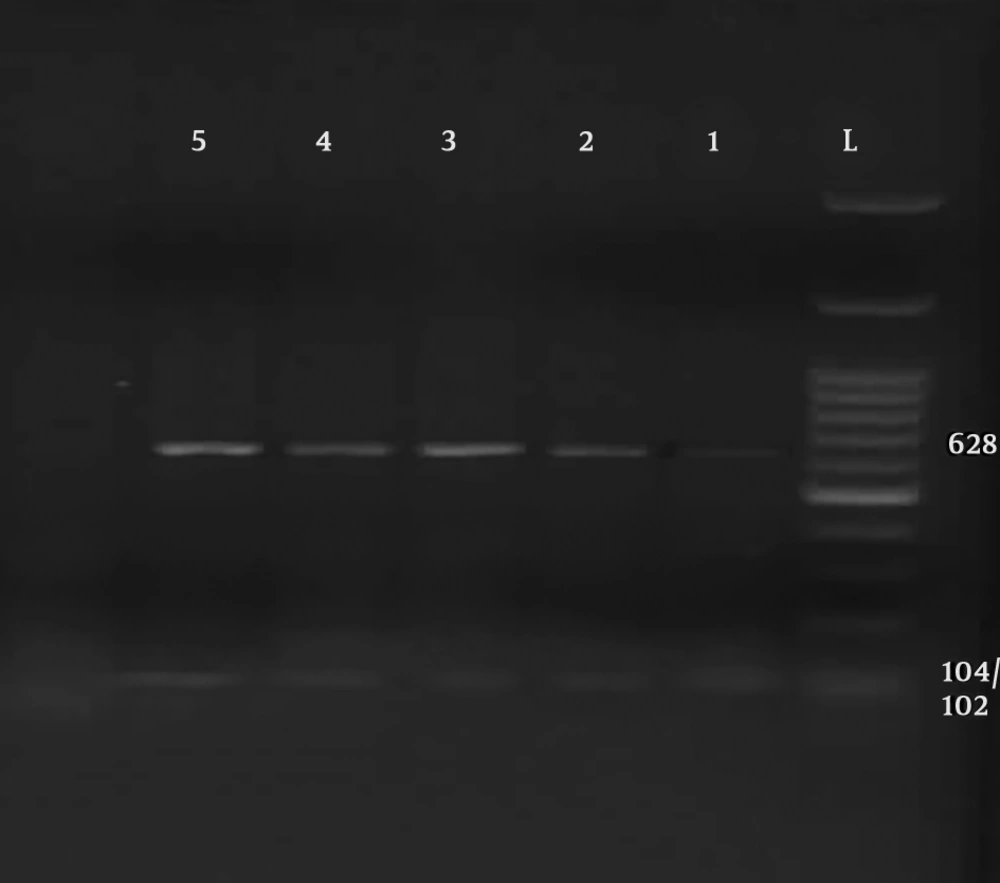
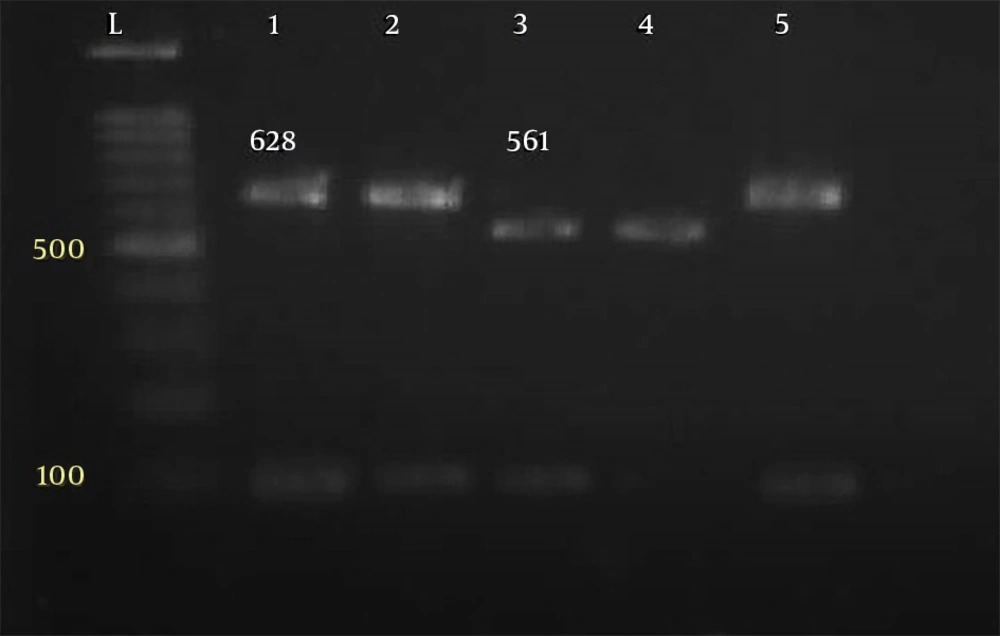
Parasitological method showed that a total of 27 out of 182 specimens were positive for Cryptosporidium oocysts. The primer used could reproduce a fragment of about 824 to 864 bp, depending on the species (Figure 1). Of the 27 human isolates examined, 17 isolates were Cryptosporidium parvum, bovine genotype, and 10 isolates belonged to Cryptosporidium hominis, human genotype (Figures 2 and 3). Table 1 shows the predicted restriction sites of the endonuclease vsp1 as it was obtained in this study.
| Species | PCR | Vsp1 Enzyme |
|---|---|---|
| C. hominis | 837 | 70,102/104,561 |
| C. parvum | 834 | 102/104,628 |
aEnzyme cutting sites with vspI by RFLP-PCR.
5. Discussion
Cryptosporidium is one of four important diarrhea pathogens in children, and a major health problems. A wide range of studies has been conducted on the various characteristics of Cryptosporidium, including biology, epidemiology, and diagnosis. The prevalence of Cryptosporidium species varies in different areas of the world. Therefore, this study was conducted to determine the species and genotype of Cryptosporidium in children with diarrhea in order to obtain more accurate information on epidemiology, control, and prevention of the parasite. In the present study, 182 samples were investigated; positive samples were selected for molecular testing. The samples later underwent PCR, using specific primers and the desired fragments were proliferated. A total of 27 samples were genotyped. A total of 17 genotypes of C. parvum and 10 isolates of C. parvum were observed. The results showed that the C. parvum bovine genotype was a dominant species and the resulting genotype pattern was consistent with those found in countries, such as France, where half of the samples belonged to bovine isolate (16), Iran (83.3%) (17), and Saudi Arabia (100%) (18). However, the other C. parvum human genotype was the dominant species in countries such as South African (81.8%) and Kenya (C. hominis 82.8%) (19).
Taghipour et al. performed molecular analysis using the Nested PCR method and the GP60 gene. He found that 89.47% and 10.52% of species belonged to C. parvum and C. hominis, respectively, and all C. parvum subtypes belonged to IId and IIa families. Mahmodpour et al. (2016) performed nested PCR using the 18SrRNA gene on patients with intestinal biopsy and observed C. parvum in three patients out of 110 patients (20). Dey et al. performed molecular analysis on immunocompromised patients using the qPCR method. They found 50.17%, 19.71%, and 2.71% of infections were caused by C. hominis, C. parvum, and both species, respectively (21). Shalaby et al. conducted a study on 100 children younger than ten years old in the city of Taif, Saudi Arabia. They aimed at investigating the prevalence and genotypes of Cryptosporidium on the 18S rRNA gene, using the PCR-RFLP method. All 11 positive species were related to C. parvum (22). Ghaffari and Kalantari performed PCR-RFLP using 18SrRNA in Iran, Malawi, Nigeria, and Vietnam, and found C. parvum, C. hominis, and C. meleagridis in 53.8%, 38.5%, and 7.7% of cases, respectively (23). In one report from Iran, isolation of Cryptosporidium spp. from human and animal hosts were characterized on the basis of both the 18S rRNA gene and Laxer locus. In this study, three Cryptosporidium species, C. hominis, C. parvum, and C. meleagridis, were recognized and similar to the current study, C. parvum was the predominant species (12).
In another study, the sequence analysis of GP60 gene showed that 17 cases (89.47%) and two cases (10.52%) belonged to C. parvum and C. hominis, respectively (20).
5.1. Conclusions
Infection with Cryptosporidium parvum is more than C. hominis in this region, and contact with livestock is considered as the most important source of human contamination. Subgenotype variation can be seen, yet dominant genotype digested with Alu1 was IIb and with Rsa1 was Ie.
5.2. Limitations of the Study
Some DNA samples were not completely extracted or disappeared during the producer, and due to emigration or cure they were missed.