Abstract
Background:
Cyclin-dependent kinase inhibitors (CKIs) are the negative regulator of cell cycle progression, which inhibits cyclin-cdk complexes, resulting in cell cycle arrest. Recently, we evaluated the effect of 5-Aza-CdR on DNMT1 gene expression in the WCH-17 hepatocellular carcinoma (HCC) cell line.Objectives:
The current study was designed to analyze the effects of 5-aza-2'–deoxycytidine (5-Aza-CdR, decitabine), 5-azacytidine (5-AzaC, vidaza), and 5'-fluoro-2'-deoxycytidine (FdCyd) on INK4a/ARF, CIP/KIP, and DNA methyltransferase 1 gene expression, apoptosis induction, and cell growth inhibition in colon cancer HCT-116 cell line.Methods:
The colon cancer HCT-116 cell line was treated with 5-azaC, 5-Aza-CdR, and FdCyd at 24 and 48h. To determine colon cancer HCT-116 cell viability, cell apoptosis, and the relative expression level of the INK4a/ARF, CIP/KIP, and DNA methyltransferase 1 genes, MTT assay, flow cytometry, and qRT-PCR were done, respectively.Results:
5-azaC, 5-Aza-CdR, and FdCyd significantly inhibited colon cancer HCT-116 cell growth and induced apoptosis. Besides, they significantly increased CIP/KIP (p21CIP1, p27KIP1, and p57KIP2) and INK4 (p14ARF, p15INK4b, and p16INK4a) and decreased DNMT1 gene expression. Besides, minimal and maximal apoptosis were seen in the groups treated with FdCyd and 5-Aza-CdR, respectively. The IC50 for CAF for FdCyd was 1.72 ± 0.23 and 1.63 ± 0.21μM at 24 and 48h, respectively. The IC50 for CAF for 5-AzaC was 2.18 ± 0.33 and 1.98 ± 0.29 μM at 24 and 48h, respectively. The IC50 for CAF for 5-Aza-CdR was 4.08 ± 0.61 and 3.18 ± 0.50 μM at 24 and 48h, respectively.Conclusions:
The 5-azac, 5-Aza-CdR, and FdCyd can reactivate the INK4a/ARF and CIP/KIP families through inhibition of DNMT1 activity.Keywords
Cyclin-dependent Kinases DNA (Cytosine-5-)-Methyltransferases Colonic Neoplasms
1. Background
The mammalian cell cycle is a highly ordered process and is divided into discrete phases including G1 the phase (gap 1), DNA synthesis phase (S phase), G2 phase, and mitosis phase (M phase). In this cycle, genetic information is transmitted from one cell to the next. The cycle is derived by the cyclin-dependent kinases (Cdks) and their regulatory subunit, cyclin, together, which propel the cell cycle through the various phases of the cell cycle (1). The transition between each phase is controlled by the kinase activity composed of cyclins and their partner, Cdk. Cyclins have fluctuated levels during the cell cycle. In contrast, the level of Cdk proteins typically remains unchanged throughout the cycle (2).
Cyclin-dependent kinase inhibitors (CKIs) are the negative regulator of cell cycle progression, which inhibit cyclin-cdk complexes, resulting in cell cycle arrest (3). Two families of CKIs have been identified: CIP/KIP and INK4 families. The first family members comprise p21CIP1 (p21), p27KIP1 (p27), and p57KIP2 (p57) (4). The INK4a/ARF includes 3 important tumor suppressor genes (TSGs): p14ARF (p14), p15INK4b (p15), and p16INK4a (p16) (5). The post-translational modifications of cell cycle include phosphorylation, histone acetylation, DNA methylation, ADP-ribosylation, and ubiquitination (6).
Aberrant DNA methylation is known as an important epigenetic change occurring in cancer. The silencing of the INK4a/ARF (7) and CIP/KIP (8) families by methylation plays an important role in several cancers. The hypermethylation of INK4a/ARF has been reported in breast cancer (9), Mantle cell lymphoma (MCL) (10), colorectal cancer (11), colon cancer (12), and gastric cancer (13). Similarly, the hypermethylation of the CIP/KIP family has been shown in the lung, gastric, and colon cancer (14). In mammals, cytosine methylation is achieved by 3 DNA methyltransferases (DNMT's) comprising DNMT1, DNMT3A, and DNMT3B (15). Several studies have indicated the role of DNMT1 in the regulation of the expression of TSGs in colon cancer cells (16).
DNA methyltransferase inhibitors (DNMTIs) can inhibit DNMTs activity resulting in the re-activation of silenced TSGs. These compounds are divided into two groups including the nucleoside inhibitors and the nonnucleoside inhibitors. First group includes 5'-fluoro-2'-deoxycytidine (FdCyd), 5-azacytidine (5-AzaC, vidaza), and 5-aza-2'–deoxycytidine (5-Aza-CdR, decitabine). The second group includes epigallocatechin3-gallate (EGCG), procaine, and RG108 (17). The inhibitory effect of these compounds has been demonstrated in lymphoid cancer, ovarian cancer, cervical cancer, lung cancer (18), breast cancer (19), and human colon cancer cells (20, 21). Recently, we evaluated the effect of 5-Aza-CdR on DNMT1 gene expression in the WCH-17 hepatocellular carcinoma (HCC) cell line (22).
2. Objectives
The current study was designed to analyze the effects of 5-azac, 5-Aza-CdR, and FdCyd on the INK4a/ARF family (p15INK4a, p14, and p15), CIP/KIP family (p21, p27, and p57), and DNA methyltransferase 1 gene expression, cell growth inhibition, and cell apoptosis induction in colon cancer HCT-116 cell line.
3. Methods
Human colon cancer HCT-116 cell line was provided from the National Cell Bank of Iran-Pasteur Institute and maintained in DMEM supplemented with fetal bovine serum 10% and antibiotics (0.1 mg/mL streptomycin and 100 U/mL penicillin). 5-azac, 5-Aza-CdR, and FdCyd were purchased from Sigma (St. Louis, MO, USA) and dissolved in DMSOto make a master stock solution (23). Further concentration was obtained by diluting the provided solution. Other compounds including materials and kits were purchased as provided for our previous works (22, 24, 25), including FBS (fetal bovine serum), MTT, Real-time PCR kits (qPCRMaster Mix Plus for SYBR Green I dNTP), and total RNA extraction Kit (TRIZOL reagent). This work is a lab trial study approved by the Ethics Committee of Jahrom University of Medical Sciences with a code number of IR.JUMS.REC.1398.099.
3.1. Cell Culture and Cell Viability
The HCT-116 cells were cultured in DMEM supplemented with 10% FBS and antibiotics (0.1 mg/ml streptomycin and 100 U/mL penicillin) at 37°C in 5% CO2 for 24 h. Subsequently, the cells were seeded into 96-well plates (4 × 105 cells per well). After 1 day, culture medium was removed and the experimental medium containing various doses of 5-azac (0, 0.5, 1, 2.5, 5, and 10 μM), 5-Aza-CdR (0, 0.5, 1, 2.5, 5, and 10 μM), and FdCyd (0, 0.5, 1, 2.5, 5, and 10 μM). The concentrations were selected based on and in the range of our previous works and other researchers' reports. The control groups were treated with the solvent (DMSO) only at a concentration of 0.05 %. After 24 and 48 h of treatment, the treated and untreated HCT-116 cells were evaluated by MTT assay to obtain cell viability, the MTT solution (5 mg/mL) was added to each well and allowed incubation for 4 h at 37°C. To dissolve all of the crystals, the solution was replaced by DMSO for 10 min. Subsequently, the absorbance spectrum was determined by a microplate reader at a wavelength of 570 nM.
3.2. Cell Apoptosis Assay
To determine cell apoptosis, the HCT-116 cells were cultured at a density of 4 × 105 cells/well and incubated overnight and, then, the cells were treated with 5-azac, 5-Aza-CdR, and FdCyd, based on IC50 values indicated in Table 1 for different periods (24 and 48 h). Subsequently, the treated and untreated HCT-116 cells were harvested and prepared for flow cytometry by trypsinization, washing twice with cold PBS, and then stained with annexin-V-(FITC) and propidium iodide (PI). The apoptotic cells were determined by FACScan flow cytometry.
IC50 Values and Apoptosis. The Cells Were Treated with Compounds, to Determine IC50 Values and Apoptosis
| Cell Line | Duration/Hour | IC50 Value/μM | LogIC50 | R Squared | Apoptosis (%) | P-Value a |
|---|---|---|---|---|---|---|
| FdCyd | 24 | 1.72 | 0.2357 | 0.7815 | 7.64 | < 0.0003 |
| FdCyd | 48 | 1.63 | 0.2121 | 0.9729 | 11.15 | < 0.0001 |
| 5-AzaC | 24 | 2.18 | 0.3391 | 0.8771 | 9.45 | < 0.0001 |
| 5-AzaC | 48 | 1.98 | 0.2981 | 0.8066 | 50.1 | < 0.0001 |
| 5-Aza-CdR | 24 | 4.08 | 0.6116 | 0.7221 | 13.16 | < 0.0001 |
| 5-Aza-CdR | 48 | 3.18 | 0.5034 | 0.7001 | 83.66 | < 0.0001 |
3.3. Real-time Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR)
To determine the relative expression level of the INK4a/ARF family (p15INK4a, p14, and p15), CIP/KIP family (p21, p27, and p57), and DNA methyltransferase 1 gene, qRT-PCR was done. The HCT-116 cells were treated with 5-azaC, 5-Aza-CdR, and FdCyd, based on IC50 values indicated in Table 1 for different periods (24 and 48 h). Total RNA from the HCT-116 cells was extracted, using the RNeasy kit according to the protocol and treated by RNase‑free DNase to eliminate the genomic DNA before cDNA synthesis (26). Real-time PCR reactions were performed, using the Steponeplus. Thermal cycling conditions were initial denaturation at 95°C for 10 minutes, followed by 40 cycles of denaturation at 95°C for 20 seconds, annealing at 58°C for 15 seconds, and extension at 72°C for 15 seconds. Thermal cycling conditions for DNMT1 was initial denaturation at 95°C for 10 minutes, followed by 40 cycles of denaturation at 95°C for 15 seconds, annealing at 60°C for 20 seconds, and extension at 72°C for 20 seconds.
The data were analyzed, using the comparative Ct (ΔΔct) method. A melting curve was used to determine the melting temperature of specific amplification products and primer dimmers. GAPDH was used as a reference gene for internal control. The primer sequences of the genes used in the current article are shown in Table 2 (27-32). The relative RT-PCR determines the expression level in comparison with a reference sample. It is based on the expression levels of a target gene versus a housekeeping gene (Pfaffl 2004) (33).
The Primer Sequences of INK4a/ARF, CIP/KIP, and DNMT1 Genes a
| Primer | Primer Sequences (5' to 3') | Product Length/bp | Reference |
|---|---|---|---|
| P14 | Forward: TACTGAGGAGCCAGCGTCTA | 146 | (27) |
| Reverse: TGCACGGGTCGGGTGAGAGT | |||
| P15 | Forward: AAGCTGAGCCCAGGTCTCCTA | 93 | (28) |
| Reverse: CCACCGTTGGCCGTAAACT | |||
| P15 | Forward: CTTCCTGGACACGCTGGT | 162 | (29) |
| Reverse: GCATGGTTACTGCCTCTGGT | |||
| P21 | Forward: CGATGGAACTTCGACTTTGTCA | 220 | (30) |
| Reverse: GCACAAGGGTACAAGACAGTG | |||
| P 27 | Forward: GGTTAGCGGAGCAATGCG | 127 | (30) |
| Reverse: TCCACAGAACCGGCATTTG | |||
| P 57 | Forward: GCGGCGATCAAGAAGCTGT | 52 | (31) |
| Reverse: GCTTGGCGAAGAAATCGGAGA | |||
| DNMT1 | Forward: GCACAAACTGACCTGCTTCA | 213 | (32) |
| Reverse: GCCTTTTCACCTCCATCAAA | |||
| GAPDH | Forward: TGTGGGCATCAATGGATTTGG | 116 | (31) |
| Reverse: ACACCATGTATTCCGGGTCAAT |
3.4. Statistical Analysis
Data from three independent experiments were analyzed with one-way analysis of variance (ANOVA) using Graphpad Prism Software version 8.0. A significant difference is expressed as P < 0.05.
4. Results
4.1. Cell Viability
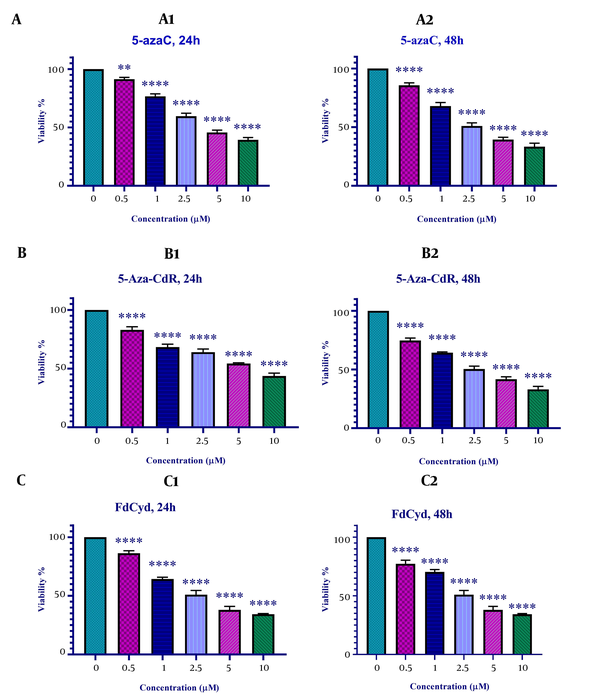
The viability of colon cancer HCT-116 cells treated with 5-azac, 5-Aza-CdR, and FdCyd was measured by MTT assay. As indicated in Figure 1, 5-azac, 5-Aza-CdR, and FdCyd induced significant cell growth inhibition. The half-maximal inhibitory concentration (IC50) values are demonstrated in Table 1. As indicated in Table 1, The IC50 for CAF for FdCyd was 1.72 ± 0.23 and 1.63 ± 0.21 μM at 24 and 48h, respectively. The IC50 for CAF for 5-AzaC was 2.18 ± 0.33 and 1.98 ± 0.29 μM at 24 and 48h, respectively. The IC50 for CAF for 5-Aza-CdR was 4.08 ± 0.61 and 3.18 ± 0.50 μM at 24 and 48h, respectively.
In vitro effects of 5-azaC, 5-Aza-CdR, and FdCyd on HCT-116 cell line viability determined by MTT Assay at 24 and 48 h. Asterisks demonstrate significant differences between HCT-116 treated and untreated control groups. It should be noted that **and **** indicate P < 0.0013 and P < 0.0001, respectively.
4.2. Cell Apoptosis
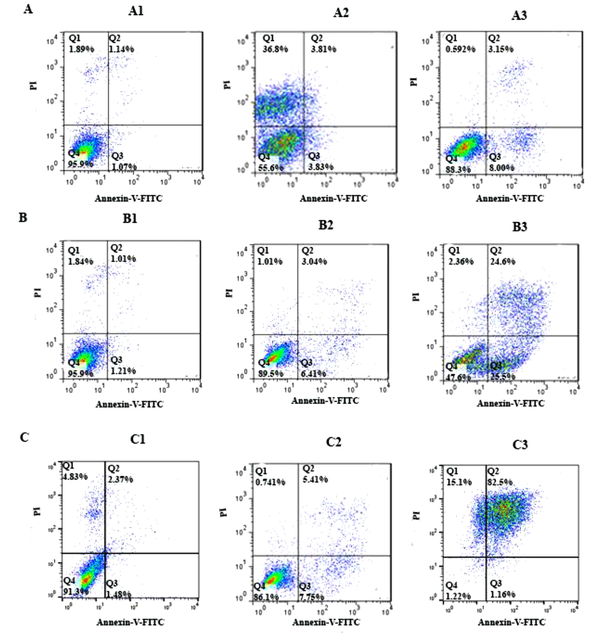
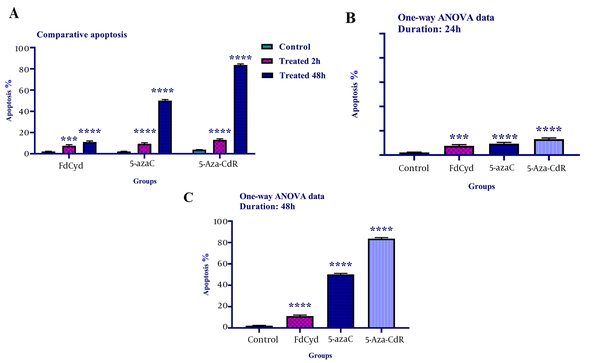
To determine cell apoptosis, the HCT-116 cells were treated with FdCyd (1.72 ± 0.23 and 1.63 ± 0.21μM), 5-AzaC (2.18 ± 0.33 and 1.98 ± 0.29 μM), and 5-Aza-CdR (4.08 ± 0.61 and 3.18 ± 0.50 μM) for 24 and 48 h and stained using annexin-V-(FITC) and PI as mentioned in the method section. As depicted in Figure 2, all of the compounds significantly induced apoptosis (Figure 3). Besides, minimal and maximal apoptosis were seen in the groups treated with FdCyd and 5-Aza-CdR, respectively. The percentage of HCT-116 apoptotic cells is shown in Table 1.
The apoptotic effect of 5-azac, 5-Aza-CdR, and FdCyd on HCT-116 cell line versus control groups at different periods (24 and 48h). The cells were treated with FdCyd (1.72 and 1.63 μM), 5-AzaC (2.18 and 1.98 μM), and 5-Aza-CdR (4.08 and 3.18 μM) for 24 and 48h, respectively. Quadrant (Q) 2 and 3, late and primary apoptosis, respectively, were calculated in this graph. A: FdCyd treated groups (A1: Control; A2: 24h; A3: 48h); B: 5-azac treated groups (B1: Control; B2: 24h; B3: 48h); C: 5-Aza-CdR treated groups (C1: Control; C2: 24h; C3: 48h).
The comparative apoptotic effects of 5-azac, 5-Aza-CdR, and FdCyd on HCT-116 cell line. The cells were treated with FdCyd (1.72 and 1.63 μM), 5-AzaC (2.18 and 1.98 μM), and 5-Aza-CdR (4.08 and 3.18 μM) for 24 and 48h, respectively. Asterisks indicate significant differences between the HCT-116 treated and untreated control groups. All compounds induced significant apoptosis in HCT-116 cell line (part A). Minimal and maximal apoptosis were seen in the groups treated with FdCyd and 5-Aza-CdR, respectively (parts B, and C). It should be noted that *** and **** indicate P < 0.0003 and P < 0.0001, respectively.
4.3. Gene Expression
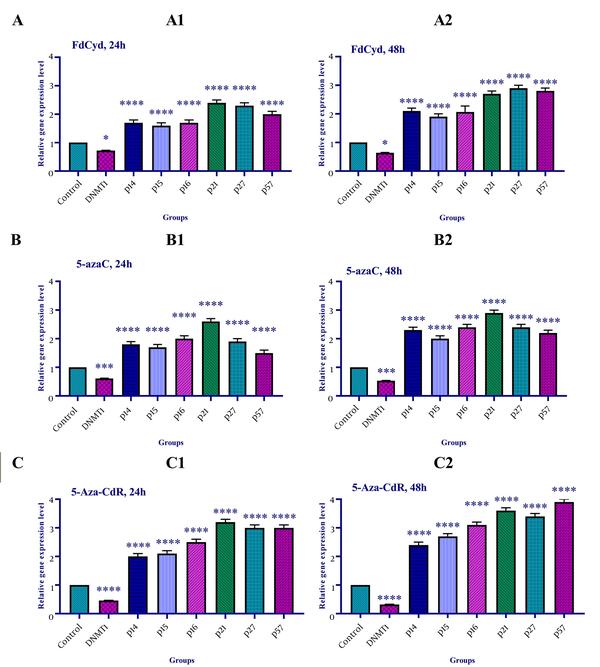
The effect of 5-azac, 5-Aza-CdR, and FdCyd on INK4a/ARF family (p15INK4a, p14, and p15), CIP/KIP family (p21, p27, and p57), and DNA methyltransferase 1 gene expression was assessed by qRT-PCR. To determine the relative gene expression, the HCT-116 cells were treated with FdCyd (1.72 ± 0.23 and 1.63 ± 0.21 μM), 5-AzaC (2.18 ± 0.33 and 1.98 ± 0.29 μM), and 5-Aza-CdR (4.08 ± 0.61 and 3.18 ± 0.50 μM) for 24 and 48h, respectively. The result indicated that treatment with 5-azac, 5-Aza-CdR, and FdCyd (24 and 48h) upregulated INK4a/ARF (p15INK4a, p14, and p15) and CIP/KIP (p21, p27, and p57) families, and down-regulated DNMT1 gene expression significantly (Figure 4).
The relative expression level of the INK4a/ARF family (p15INK4a, p14, and p15), CIP/KIP family (p21, p27, and p57), and DNA methyltransferase 1 gene in the colon cancer HCT-116 cell line. The cells were treated with FdCyd (1.72 and 1.63 μM), 5-AzaC (2.18 and 1.98 μM), and 5-Aza-CdR (4.08 and 3.18 μM) for 24 and 48h, respectively. A significant difference was seen between treated and untreated control groups. Asterisks indicate significant differences between the treated and untreated control groups. A: FdCyd treated groups; B: 5-azac treated groups; C: 5-Aza-CdR treated groups. It should be noted that *, ***, and **** indicate P < 0.0195, P < 0.0011, and P < 0.0001, respectively.
5. Discussion
Cellular gene transcription is directly under the influence of the genomic structure and organization. DNA methylation is an important epigenetic modification that has widespread influences on gene transcription and expression (34). It is profoundly altered in human cancers. The silencing of TSGs by this epigenetic change is known as a key mechanism in tumorigenesis (35). The potential anticancer activity of DNMT inhibitors has been extensively evaluated in recent years. These compounds are widely studied because DNA demethylation induces the re-activation of TSGs that are silenced by promoter hypermethylation (36).
In the present study, we demonstrated that DNA demethylating agents 5-azac, 5-Aza-CdR, and FdCyd inhibited HCT-116 cell growth and induced apoptosis in colon cancer HCT-116 cell lines. Furthermore, we did a further evaluation to find the molecular mechanism of the compounds. Thus, we found that these agents could re-activate INK4a/ARF family (p15INK4a, p14, and p15) and the CIP/KIP family (p21, p27, and p57) by inhibition of DNA methyltransferase 1 gene expression.
Similar apoptotic pathways have been shown by other researchers. It has been indicated that 5-Aza-CdR increases the expression of both p15INK4a and p19INK4d in the human lung cancer cell line (37). In vitro studies have shown such molecular mechanisms for the member of INK4 in colorectal cancer (38), leukemia (39), ovarian cancer (40), gastric cancer (41), and HCC (42). As mentioned above DNA methyltransferase inhibitors play their role through the re-activation of the CIP/KIP family. A similar pathway has been indicated in HCC (43), gastric cancer (44), breast, and lung cancer (45). We reported that DNA demethylating agents 5-azac, 5-Aza-CdR, and FdCyd play their role through inhibition of DNMT1 activity.
In addition to this function, several in vitro studies have been demonstrated that these agents can inhibit DNMT3a and DNMT3b in the colon cancer HCT-116 (46), DNMT3B in human endometrial cancer (47), DNMT1, and/or DNAT 3b mediates in lung cancer, esophageal cancer, and malignant pleural mesothelioma cells (48), and DNMT3b in testicular germ cell tumors (TGCT) (49).
Additionally, other members of DNA methyltransferase inhibitors can induce apoptosis with similar molecular mechanisms. Our previous work indicated that zebularine (a membership of HDACIs) can induce apoptosis through down-regulation of DNMT1, DNMT3a, and DNMT3b and up-regulation of p21Cip1/Waf1/Sdi1, p27Kip1, and p57Kip2 in colon cancer LS 174T cell line (50). Besides, we reported that zebularine induces apoptosis by down-regulation of DNMT1, 3a, and 3b and up-regulation of p21Cip1/Waf1/Sdi1, p27Kip1, p57Kip2 in colon cancer LS 180 cell line (51). Meanwhile, DNMT1 inhibition is not the only apoptotic pathway of DNMTIs. It has been reported that 5 aza 2' deoxycytidine treatment resulted in significant FAS gene up-regulation in the HT 29 cell line and it plays its role through the extrinsic apoptotic pathway (52). Inconsistent with our report, it has been reported that 5 AZA treatments cannot induce a significant change in cell proliferation, cell cycle arrest, cell apoptosis, and mtDNA copy number in HCT116, SW480, LS 174T, and HT 29 cell lines (53). Finally, DNMTIs can play their apoptotic roles through various molecular mechanisms.
We indicated that inhibition of DNMT1 activity by 5-azac, 5-Aza-CdR, and FdCyd induces re-activation of INK4a/ARF family (p15INK4a, p14, and p15) and CIP/KIP family (p21, p27, and p57), resulting in apoptosis induction in colon cancer HCT-116 cell line.
We did not assess the other DNMTs gene expression such as DNMT3a and DNMT3b in this work. Thus, this evaluation is recommended.
5.1. Conclusions
In summary, our findings indicated that 5-azac, 5-Aza-CdR, and FdCyd inhibited colon cancer HCT-116 cell line and induced apoptosis in this cell line. The most likely molecular mechanism underlying these compounds inhibited HCT-116 cell growth and induced apoptosis involves down-regulation of DNMT1 and up-regulation of CIP/KIP (p21, p27, and p57) and INK4 (p14, p15, and p15INK4a) genes expression. This result suggests that 5-azac, 5-Aza-CdR, and FdCyd may have wide therapeutic applications in colon cancer.
References
-
1.
Israels ED, Israels LG. The cell cycle. Oncologist. 2000;5(6):510-3. [PubMed ID: 11110604]. https://doi.org/10.1634/theoncologist.5-6-510.
-
2.
Lim S, Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140(15):3079-93. [PubMed ID: 23861057]. https://doi.org/10.1242/dev.091744.
-
3.
Shankland SJ, Wolf G. Cell cycle regulatory proteins in renal disease: role in hypertrophy, proliferation, and apoptosis. Am J Physiol Renal Physiol. 2000;278(4):F515-29. [PubMed ID: 10751212]. https://doi.org/10.1152/ajprenal.2000.278.4.F515.
-
4.
Funk JO. Cell cycle checkpoint genes and cancer. 5. Wiley; 2001. p. 1-6.
-
5.
Zhang Z, Rosen DG, Yao JL, Huang J, Liu J. Expression of p14ARF, p15INK4b, p16INK4a, and DCR2 increases during prostate cancer progression. Mod Pathol. 2006;19(10):1339-43. [PubMed ID: 16799475]. https://doi.org/10.1038/modpathol.3800655.
-
6.
Shanmugam MK, Arfuso F, Arumugam S, Chinnathambi A, Jinsong B, Warrier S, et al. Role of novel histone modifications in cancer. Oncotarget. 2018;9(13):11414-26. [PubMed ID: 29541423]. [PubMed Central ID: PMC5834259]. https://doi.org/10.18632/oncotarget.23356.
-
7.
Park MT, Lee SJ. Cell cycle and cancer. J Biochem Mol Biol. 2003;36(1):60-5. [PubMed ID: 12542976]. https://doi.org/10.5483/bmbrep.2003.36.1.060.
-
8.
Chim CS, Fung TK, Wong KF, Lau JS, Law M, Liang R. Methylation of INK4 and CIP/KIP families of cyclin-dependent kinase inhibitor in chronic lymphocytic leukaemia in Chinese patients. J Clin Pathol. 2006;59(9):921-6. [PubMed ID: 16565223]. [PubMed Central ID: PMC1860467]. https://doi.org/10.1136/jcp.2005.035089.
-
9.
Rivandi M, Khorrami MS, Fiuji H, Shahidsales S, Hasanzadeh M, Jazayeri MH, et al. The 9p21 locus: A potential therapeutic target and prognostic marker in breast cancer. J Cell Physiol. 2018;233(7):5170-9. [PubMed ID: 29240242]. https://doi.org/10.1002/jcp.26332.
-
10.
Hutter G, Scheubner M, Zimmermann Y, Kalla J, Katzenberger T, Hubler K, et al. Differential effect of epigenetic alterations and genomic deletions of CDK inhibitors [p16(INK4a), p15(INK4b), p14(ARF)] in mantle cell lymphoma. Genes Chromosomes Cancer. 2006;45(2):203-10. [PubMed ID: 16258956]. https://doi.org/10.1002/gcc.20277.
-
11.
Ishiguro A, Takahata T, Saito M, Yoshiya G, Tamura Y, Sasaki M, et al. Influence of methylated p15 and p16 genes on clinicopathological features in colorectal cancer. J Gastroenterol Hepatol. 2006;21(8):1334-9. [PubMed ID: 16872319]. https://doi.org/10.1111/j.1440-1746.2006.04137.x.
-
12.
Burri N, Shaw P, Bouzourene H, Sordat I, Sordat B, Gillet M, et al. Methylation silencing and mutations of the p14ARF and p16INK4a genes in colon cancer. Lab Invest. 2001;81(2):217-29. [PubMed ID: 11232644]. https://doi.org/10.1038/labinvest.3780230.
-
13.
Zhang F, Wang L, Wu PP, Yan ZW, Zheng L, Yu YY, et al. In situ analysis of p16/INK4 promoter hypermethylation in esophageal carcinoma and gastric carcinoma. Chin J Dig Dis. 2004;5(4):149-55. [PubMed ID: 15612883]. https://doi.org/10.1111/j.1443-9573.2004.00172.x.
-
14.
Fang JY, Lu YY. Effects of histone acetylation and DNA methylation on p21( WAF1) regulation. World J Gastroenterol. 2002;8(3):400-5. [PubMed ID: 12046058]. [PubMed Central ID: PMC4656409]. https://doi.org/10.3748/wjg.v8.i3.400.
-
15.
Robertson KD. DNA methylation, methyltransferases, and cancer. Oncogene. 2001;20(24):3139-55. [PubMed ID: 11420731]. https://doi.org/10.1038/sj.onc.1204341.
-
16.
Fang JY, Lu R, Mikovits JA, Cheng ZH, Zhu HY, Chen YX. Regulation of hMSH2 and hMLH1 expression in the human colon cancer cell line SW1116 by DNA methyltransferase 1. Cancer Lett. 2006;233(1):124-30. [PubMed ID: 16473673]. https://doi.org/10.1016/j.canlet.2005.03.005.
-
17.
Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005;97(20):1498-506. [PubMed ID: 16234563]. https://doi.org/10.1093/jnci/dji311.
-
18.
Gnyszka A, Jastrzebski Z, Flis S. DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res. 2013;33(8):2989-96. [PubMed ID: 23898051].
-
19.
Billam M, Sobolewski MD, Davidson NE. Effects of a novel DNA methyltransferase inhibitor zebularine on human breast cancer cells. Breast Cancer Res Treat. 2010;120(3):581-92. [PubMed ID: 19459041]. [PubMed Central ID: PMC3901992]. https://doi.org/10.1007/s10549-009-0420-3.
-
20.
Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, et al. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33(1):61-5. [PubMed ID: 12496760]. https://doi.org/10.1038/ng1068.
-
21.
Thakur S, Feng X, Qiao Shi Z, Ganapathy A, Kumar Mishra M, Atadja P, et al. ING1 and 5-azacytidine act synergistically to block breast cancer cell growth. PLoS One. 2012;7(8). e43671. [PubMed ID: 22916295]. [PubMed Central ID: PMC3423394]. https://doi.org/10.1371/journal.pone.0043671.
-
22.
Sanaei M, Kavoosi F. Effects of 5-aza-2'-deoxycytidine and Valproic Acid on Epigenetic-modifying DNMT1 Gene Expression, Apoptosis Induction and Cell Viability in Hepatocellular Carcinoma WCH-17 cell line. Iran J Pediatr Hematol Oncol. 2019;9(2):83-90. https://doi.org/10.18502/ijpho.v9i2.607.
-
23.
Sanaei M, Kavoosi F. Effect of 5-Aza-2'-Deoxycytidine in Comparison to Valproic Acid and Trichostatin A on Histone Deacetylase 1, DNA Methyltransferase 1, and CIP/KIP Family (p21, p27, and p57) Genes Expression, Cell Growth Inhibition, and Apoptosis Induction in Colon Cancer SW480 Cell Line. Adv Biomed Res. 2019;8:52. [PubMed ID: 31516890]. [PubMed Central ID: PMC6712896]. https://doi.org/10.4103/abr.abr_91_19.
-
24.
Sanaei M, Kavoosi F. Effect of DNA Methyltransferase in Comparison to and in Combination with Histone Deacetylase Inhibitors on Hepatocellular Carcinoma HepG2 Cell Line. Asian Pac J Cancer Prev. 2019;20(4):1119-25. [PubMed ID: 31030484]. [PubMed Central ID: PMC6948907]. https://doi.org/10.31557/APJCP.2019.20.4.1119.
-
25.
Kavoosi F, Sanaei M. Comparative Analysis of the Effects of Valproic Acid and Tamoxifen on Proliferation, and Apoptosis of Human Hepatocellular Carcinoma WCH 17 CellLin. Iran J Pediatr Hematol Oncol. 2018;8(1):12-20.
-
26.
Sanaei M, Kavoosi F. Effect of Curcumin and Trichostatin A on the Expression of DNA Methyltransfrase 1 in Hepatocellular Carcinoma Cell Line Hepa 1-6. Iran J Pediatr Hematol Oncol. 2018;8(4):193-201.
-
27.
Maruo S, Zhao B, Johannsen E, Kieff E, Zou J, Takada K. Epstein-Barr virus nuclear antigens 3C and 3A maintain lymphoblastoid cell growth by repressing p16INK4A and p14ARF expression. Proc Natl Acad Sci U S A. 2011;108(5):1919-24. [PubMed ID: 21245331]. [PubMed Central ID: PMC3033265]. https://doi.org/10.1073/pnas.1019599108.
-
28.
Li G, Ji Y, Liu C, Li J, Zhou Y. Reduced levels of p15INK4b, p16INK4a, p21cip1 and p27kip1 in pancreatic carcinoma. Mol Med Rep. 2012;5(4):1106-10. [PubMed ID: 22293850]. [PubMed Central ID: PMC3493078]. https://doi.org/10.3892/mmr.2012.771.
-
29.
Wang Y, Zang X, Wang Y, Chen P. High expression of p16INK4a and low expression of Bmi1 are associated with endothelial cellular senescence in the human cornea. Mol Vis. 2012;18:803-15. [PubMed ID: 22509111]. [PubMed Central ID: PMC3324359].
-
30.
Wang L, Wang G, Yang D, Guo X, Xu Y, Feng B, et al. Euphol arrests breast cancer cells at the G1 phase through the modulation of cyclin D1, p21 and p27 expression. Mol Med Rep. 2013;8(4):1279-85. [PubMed ID: 23969579]. https://doi.org/10.3892/mmr.2013.1650.
-
31.
Xu Y, Zhong C, Ding S, Huang H, Shen Z. MicroRNA-221 promotes human non-small cell lung cancer cell H460 growth. Int J Clin Exp Med. 2015;8(2):2024-30. [PubMed ID: 25932132]. [PubMed Central ID: PMC4402779].
-
32.
Xu Y, Chao L, Wang J, Sun Y. miRNA-148a regulates the expression of the estrogen receptor through DNMT1-mediated DNA methylation in breast cancer cells. Oncol Lett. 2017;14(4):4736-40. [PubMed ID: 29085474]. [PubMed Central ID: PMC5649610]. https://doi.org/10.3892/ol.2017.6803.
-
33.
Pfaffl MW. Quantification strategies in real-time PCR. AZ of quantitative PCR. 2004;1:89-113.
-
34.
Metivier R, Gallais R, Tiffoche C, Le Peron C, Jurkowska RZ, Carmouche RP, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452(7183):45-50. [PubMed ID: 18322525]. https://doi.org/10.1038/nature06544.
-
35.
Kondo Y. Epigenetic cross-talk between DNA methylation and histone modifications in human cancers. Yonsei Med J. 2009;50(4):455-63. [PubMed ID: 19718392]. [PubMed Central ID: PMC2730606]. https://doi.org/10.3349/ymj.2009.50.4.455.
-
36.
Zhu WG, Otterson GA. The interaction of histone deacetylase inhibitors and DNA methyltransferase inhibitors in the treatment of human cancer cells. Curr Med Chem Anticancer Agents. 2003;3(3):187-99. [PubMed ID: 12769777]. https://doi.org/10.2174/1568011033482440.
-
37.
Zhu WG, Dai Z, Ding H, Srinivasan K, Hall J, Duan W, et al. Increased expression of unmethylated CDKN2D by 5-aza-2'-deoxycytidine in human lung cancer cells. Oncogene. 2001;20(53):7787-96. [PubMed ID: 11753657]. https://doi.org/10.1038/sj.onc.1204970.
-
38.
Zheng S, Chen P, McMillan A, Lafuente A, Lafuente MJ, Ballesta A, et al. Correlations of partial and extensive methylation at the p14(ARF) locus with reduced mRNA expression in colorectal cancer cell lines and clinicopathological features in primary tumors. Carcinogenesis. 2000;21(11):2057-64. [PubMed ID: 11062168]. https://doi.org/10.1093/carcin/21.11.2057.
-
39.
Farinha NJ, Shaker S, Lemaire M, Momparler L, Bernstein M, Momparler RL. Activation of expression of p15, p73 and E-cadherin in leukemic cells by different concentrations of 5-aza-2’-deoxycytidine (Decitabine). Anticancer Res. 2004;24(1):75-8.
-
40.
Meng CF, Su B, Li W. DNA demethylation is superior to histone acetylation for reactivating cancer-associated genes in ovarian cancer cells. Mol Med Rep. 2011;4(6):1273-8. [PubMed ID: 21850374]. https://doi.org/10.3892/mmr.2011.557.
-
41.
Meng CF, Zhu XJ, Peng G, Dai DQ. Promoter histone H3 lysine 9 di-methylation is associated with DNA methylation and aberrant expression of p16 in gastric cancer cells. Oncol Rep. 2009;22(5):1221-7. [PubMed ID: 19787243]. https://doi.org/10.3892/or_00000558.
-
42.
Liu LH, Xiao WH, Liu WW. Effect of 5-Aza-2'-deoxycytidine on the P16 tumor suppressor gene in hepatocellular carcinoma cell line HepG2. World J Gastroenterol. 2001;7(1):131-5. [PubMed ID: 11819749]. [PubMed Central ID: PMC4688690]. https://doi.org/10.3748/wjg.v7.i1.131.
-
43.
Matsuda Y. Molecular mechanism underlying the functional loss of cyclindependent kinase inhibitors p16 and p27 in hepatocellular carcinoma. World J Gastroenterol. 2008;14(11):1734-40. [PubMed ID: 18350604]. [PubMed Central ID: PMC2695913]. https://doi.org/10.3748/wjg.14.1734.
-
44.
Sun D, Toan X, Zhang Y, Chen Y, Lu R, Wang X, et al. Mammalian target of rapamycin pathway inhibition enhances the effects of 5-aza-dC on suppressing cell proliferation in human gastric cancer cell lines. Sci China C Life Sci. 2008;51(7):640-7. [PubMed ID: 18622747]. https://doi.org/10.1007/s11427-008-0080-2.
-
45.
Kobatake T, Yano M, Toyooka S, Tsukuda K, Dote H, Kikuchi T, et al. Aberrant methylation of p57KIP2 gene in lung and breast cancers and malignant mesotheliomas. Oncol Rep. 2004;12(5):1087-92. [PubMed ID: 15492797].
-
46.
Schneider-Stock R, Diab-Assef M, Rohrbeck A, Foltzer-Jourdainne C, Boltze C, Hartig R, et al. 5-Aza-cytidine is a potent inhibitor of DNA methyltransferase 3a and induces apoptosis in HCT-116 colon cancer cells via Gadd45- and p53-dependent mechanisms. J Pharmacol Exp Ther. 2005;312(2):525-36. [PubMed ID: 15547111]. https://doi.org/10.1124/jpet.104.074195.
-
47.
Cui M, Wen Z, Chen J, Yang Z, Zhang H. 5-Aza-2'-deoxycytidine is a potent inhibitor of DNA methyltransferase 3B and induces apoptosis in human endometrial cancer cell lines with the up-regulation of hMLH1. Med Oncol. 2010;27(2):278-85. [PubMed ID: 19306077]. https://doi.org/10.1007/s12032-009-9204-1.
-
48.
Kassis ES, Zhao M, Hong JA, Chen GA, Nguyen DM, Schrump DS. Depletion of DNA methyltransferase 1 and/or DNA methyltransferase 3b mediates growth arrest and apoptosis in lung and esophageal cancer and malignant pleural mesothelioma cells. J Thorac Cardiovasc Surg. 2006;131(2):298-306. [PubMed ID: 16434257]. https://doi.org/10.1016/j.jtcvs.2005.05.022.
-
49.
Beyrouthy MJ, Garner KM, Hever MP, Freemantle SJ, Eastman A, Dmitrovsky E, et al. High DNA methyltransferase 3B expression mediates 5-aza-deoxycytidine hypersensitivity in testicular germ cell tumors. Cancer Res. 2009;69(24):9360-6. [PubMed ID: 19951990]. [PubMed Central ID: PMC2795063]. https://doi.org/10.1158/0008-5472.CAN-09-1490.
-
50.
Sanaei M, Kavoosi F. Effect of Zebularine in Comparison to and in Combination with Trichostatin A on CIP/KIP Family (p21Cip1/Waf1/Sdi1, p27Kip1, and p57Kip2), DNMTs (DNMT1, DNMT3a, and DNMT3b), Class I HDACs (HDACs 1, 2, 3) and Class II HDACs (HDACs 4, 5, 6) Gene Expression, Cell Growth Inhibition and Apoptosis Induction in Colon Cancer LS 174T Cell Line. Asian Pac J Cancer Prev. 2020;21(7):2131-9. [PubMed ID: 32711442]. [PubMed Central ID: PMC7573409]. https://doi.org/10.31557/APJCP.2020.21.7.2131.
-
51.
Sanaei M, Kavoosi F. Investigation of the Effect of Zebularine in Comparison to and in Combination with Trichostatin A on p21Cip1/Waf1/ Sdi1, p27Kip1, p57Kip2, DNA Methyltransferases and Histone Deacetylases in Colon Cancer LS 180 Cell Line. Asian Pac J Cancer Prev. 2020;21(6):1819-28. [PubMed ID: 32592383]. [PubMed Central ID: PMC7568903]. https://doi.org/10.31557/APJCP.2020.21.6.1819.
-
52.
Manoochehri M, Borhani N, Karbasi A, Koochaki A, Kazemi B. Promoter hypermethylation and downregulation of the FAS gene may be involved in colorectal carcinogenesis. Oncol Lett. 2016;12(1):285-90. [PubMed ID: 27347139]. [PubMed Central ID: PMC4906593]. https://doi.org/10.3892/ol.2016.4578.
-
53.
Tong H, Zhang L, Gao J, Wen S, Zhou H, Feng S. Methylation of mitochondrial DNA displacement loop region regulates mitochondrial copy number in colorectal cancer. Mol Med Rep. 2017;16(4):5347-53. [PubMed ID: 28849075]. [PubMed Central ID: PMC5647067]. https://doi.org/10.3892/mmr.2017.7264.