1. Background
The most common cancer among women worldwide is breast cancer (BC), which is also the fifth leading cause of cancer-related death (1). Although there have been notable recent achievements in diagnosis and treatment, 20% - 30% of BC patients develop metastasis (2). One of the factors that can lead to increased cancer mortality is the metastasis of cancer cells to vital organs (3). In general, changes in the expression of some genes can stimulate metastasis in certain organs (4). Metastasis affects different sites and has different rates depending on the primary tumor subgroup (5).
BC includes almost four molecular subtypes: Luminal A, luminal B, HER2-enriched, and triple-negative (6). About 60% of all patients with BC have luminal A subtype of this disease (7, 8). HER2-enriched BC occurs in 15% - 25% of patients due to increased expression of the human epidermal growth factor receptor 2 (HER2) gene and is more aggressive (9, 10).
Time is a crucial factor in the detection of BC; therefore, some approaches are used for timely diagnosis and prognosis. Current diagnostic and therapeutic methods have some weaknesses; thus, molecular markers can be widely used in the diagnosis of BC. Accordingly, the main candidates for early-stage biomarkers are proteins, gene expression, and microRNA expression (11).
MiRNAs are short single-stranded RNAs with 18 - 23 nucleotides and are obtained from 70 nucleotide precursors. They manage both physiological and developmental processes through gene expression, and consequently, they play a notable role in gene expression and post-transcriptional regulation (12-15).
Recent studies have shown that miRNAs are recognized as important biomarkers in many cancers (16). Furthermore, each microRNA in humans can target hundreds of mRNAs; thus, they are able to regulate several cellular processes, like development, differentiation, and proliferation (17). Based on miRNA databases, there are more than 2500 mature microRNA sequences in the human genome (18, 19). It is estimated that more than 60% of the proteins encoding genes in humans in their 3’-UTRs have miRNA binding sites (20). MicroRNAs interpose the suppression of mRNAs by pairing to the complementary base sequences in 3’UTR, which causes instability in transcription or repression in translation or both (21). Some recent studies have shown that the expression of some genes is also regulated by exon-linked microRNAs (22-24).
2. Objectives
In the present study, the eligibility of miR-200c and miR-30a as biomarkers of BC was studied. Additionally, the relationships between expression levels of these microRNAs and tumor stage, HER2 expression, and lymphatic metastasis were also assessed.
3. Methods
3.1. Patients and Specimens
We included 30 patients with ductal carcinoma, of whom 18 cases were less than 50 years old. Also, 14 patients had never received hormone therapy, while the rest had received this therapy (Table 1). The normal and tumor samples were collected from the cancer institute of Imam Khomeini Hospital Affiliated with Tehran University of Medical Sciences. Before any analysis, the subjects completed the informed consent. The samples were conserved in RNase-free tubes, put immediately in liquid nitrogen, sent to the laboratory, and stored at -80°C.
| Characteristics | Valuesa |
|---|---|
| Age | |
| > 50 | 18 (60) |
| ≤ 50 | 12 (40) |
| Total | 30 (100) |
| Median | 49.32 (30 - 76) |
| Tumor size, cm | |
| < 3 | 12 (40) |
| ≤ 3 | 18 (60) |
| Metastasis to lymph nodes | |
| Positive | 11 (37) |
| Negative | 19 (63) |
| ER | 18 (60) |
| ER+ | 9 (30) |
| ER-unknown | 3 (10) |
| HER2 status | |
| Her2+ | 16 (53) |
| Her2- | 13 (43) |
| Unknown | 1 (0.3) |
aValues are expressed as No. (%) unless otherwise indicated.
3.2. RNA Extraction and Real-time PCR
Total RNA from the patient was isolated (Parsgenome RNA extraction kit), following the manufacturer’s protocol. Next, quantitative and qualitative evaluation was performed by UV spectrophotometry (via NanoDrop® ND-1000). Moreover, the miRNA cDNA was synthesized using the miRNA cDNA Synthesis kit (Parsgenome) and used as a template for a quantitative real-time PCR (qRT-PCR). Next, real-time PCR was performed on an Applied Bio-system real-time PCR instrument with microRNA and universal and specific primers. We used the U6 snRNA gene as the housekeeping gene to compare target RNA expression levels. All reactions were performed in triplicate. Afterward, the data were converted to cycle threshold (Ct), to normalize each microRNA expression ratio with 2-ΔΔCT method, where ΔCt = U6 ΔCt - miRNA ΔCt.
3.3. Melting Curve Analysis
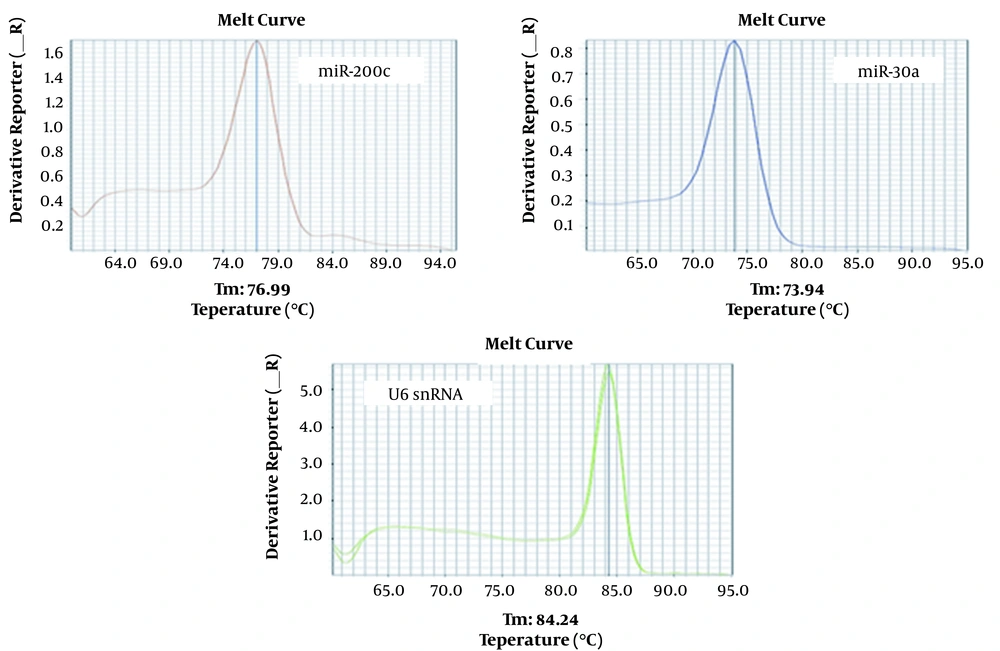
In our study, having only one peak for each gene’s melting curve showed specificity in the PCR, whereas there would have been more curves (shorter ones) if there was impurity (such as dimer primers).
3.4. Relationship Between Breast Cancer Pathogenesis and Selected microRNAs
To discover the relationship between BC pathogenesis and selected microRNAs, we measured the miR-200c and miR-30a expression levels in tumor tissue from BC patients and normal tissues surrounding it (n = 30 patients).
3.5. The Potential of miR-200c and miR-30a to Be Selected as Biomarkers
To assess the miR-200c and miR-30a potential as biomarkers for diagnosing the breast tumor tissues, we analyzed the receiver operating characteristics (ROC) curve.
3.6. Statistical Analysis
Data were expressed as the median ± standard deviation obtained from 2 or 3 separate experiments. The statistical data analysis was done using a t-test. A P-value < 0.05 was considered to be significant.
4. Results
4.1. Melting Curve Analysis and Specificity of Real-time PCR Products
Curve analysis for U6snRNA, miR-200c, and miR-30a showed only one peak for these genes at the specific melting temperature, which indicated the specificity of the PCR products (Figure 1).
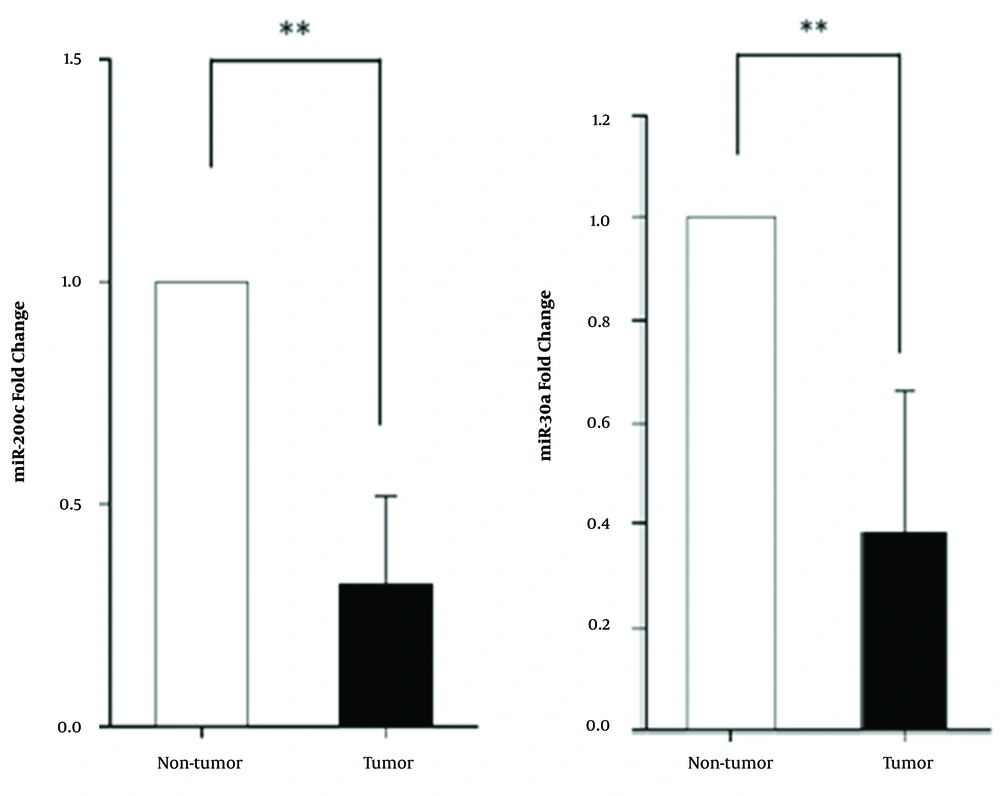
4.2. The miR-200c and miR-30a Are Significantly Down-Regulated in Tumor Tissues of Breast Cancer Patients
Our results showed a notably lower than normal expression level for miR-200c in tumor tissue compared to healthy tissue around the tumor. The miR-30a also had a lower expression level in BC patients (P < 0. 01) (Figure 2).
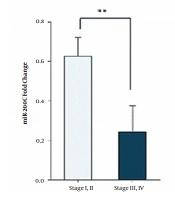
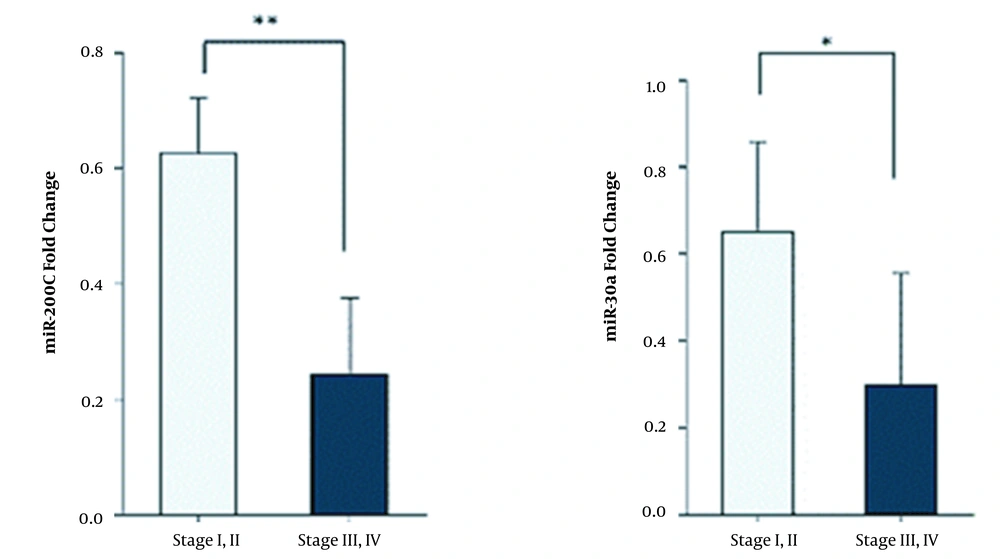
4.3. Decreased Expression Level of miR-200c and miR-30a Is Associated with Higher Stages of Breast Cancer
To observe miR-200c and miR-30a expression levels in different stages, we divided cancer stages into two groups (stage I and II versus stage III and IV). The results indicated that miR-200c and miR-30a expression levels were lower in stages III and IV (P < 0. 01) (Figure 3).
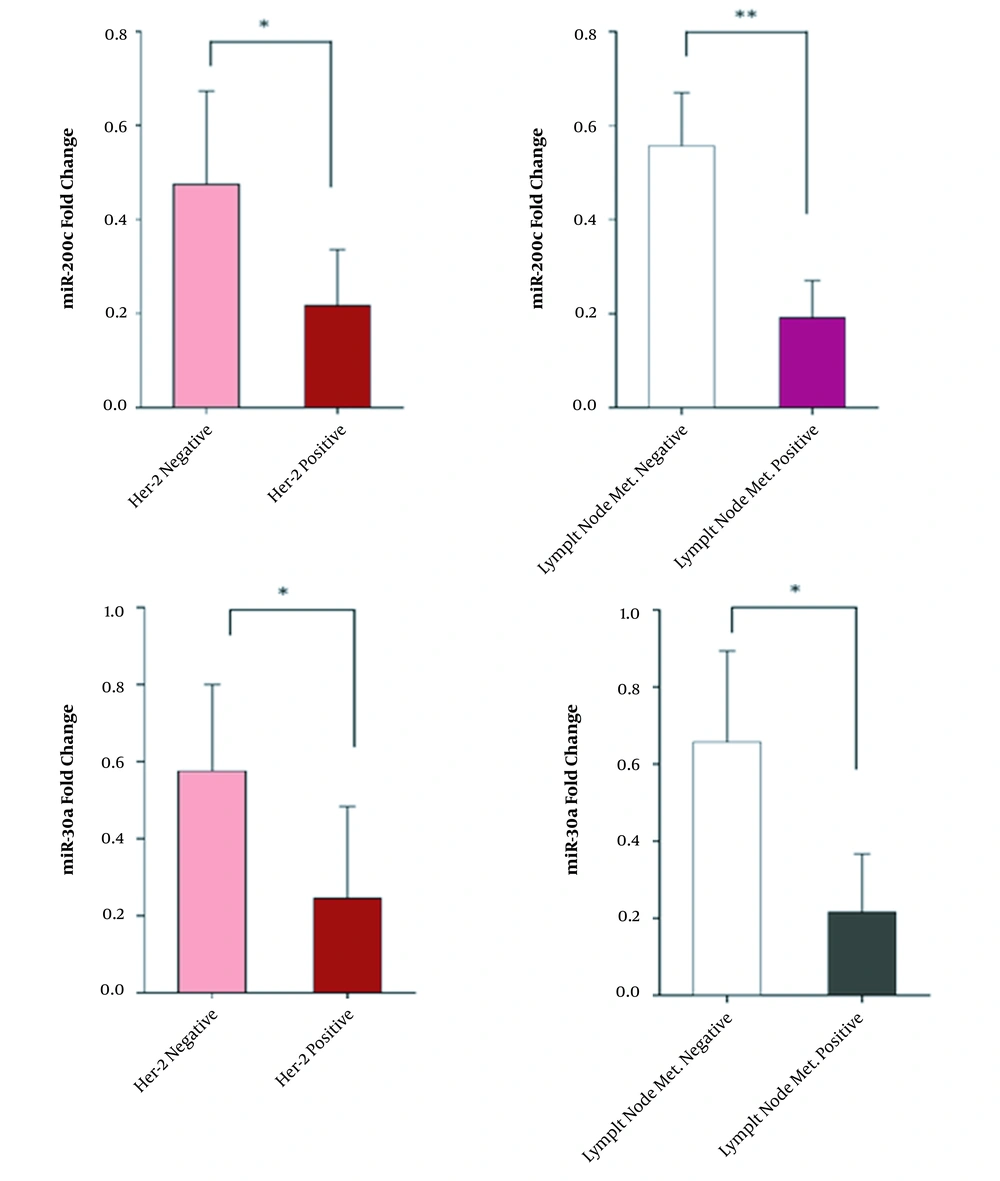
4.4. The miR-200c and miR-30a Expression Levels in HER-2-Positive Patients and Patients with Metastasis to Lymph Nodes
We also evaluated the expression levels of miR-200c and miR-30a in patients with lymph node metastasis compared to non-metastatic cases and also in HER-2-positive patients compared to HER-2-negative patients (Figure 4).
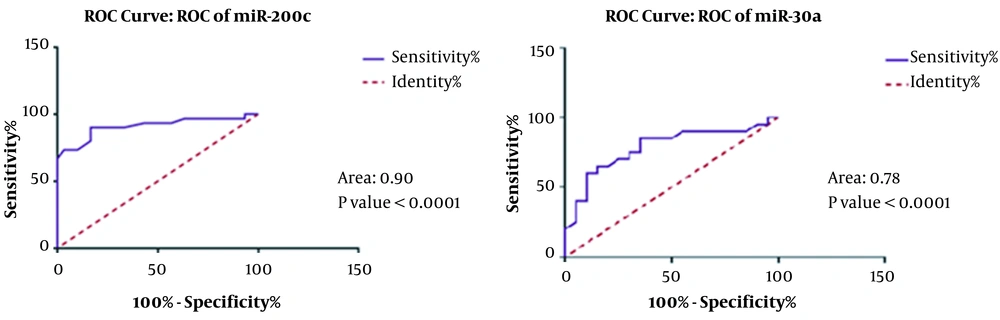
4.5. The miR-200c and miR-30a Are Appropriate Biomarkers in Breast Cancer
Our results indicated that both microRNAs showed lower expression levels in tumor tissues than normal ones. The area under the curve (AUC) indicating the potential of miR-200c and miR-30a as biomarkers was 0.90 and 0.78, respectively, which showed their high potential to be considered as biomarkers (Figure 5).
5. Discussion
The most common method for early detection of BC is mammography, which provides a high rate of false-positive results. Other methods include the use of breast imaging and biopsy, which are invasive methods that affect the quality of life. The use of sensitive methods is essential for the early detection of BC (25). MicroRNAs are important regulators that play a regulatory role in many types of cancer (26). They are stable molecules in body fluids, including blood and cerebrospinal fluid; thus, they are potent biomarkers for early non-invasive detection of BC (27).
MiR-30a is a target molecule associated with epithelial-mesenchymal transmission (EMT) that suppresses migration and invasion in some cancers, including BC (28-30). It also inhibits several important inhibitors, including Eya2, ITGB3, and UBE3c (31-34). Zeng et al. (35) showed that miR-30a levels were significantly associated with estrogen receptor (ER) and triple-negative BC (P = 0.005 and P = 0.007, respectively). In addition, they showed that the sensitivity and specificity of miR-30a for BC diagnosis were 74% and 65.6%, respectively (35). Kawaguchi et al. found that miR-30a and miR-200c had a significant prognosis in the Cancer Genome Atlas (TCGA) data portal. Their study showed that the overexpression of these tumor suppressors, including miR-30a and mir-200c, was associated with overall survival (OS) (36). Hurteau et al. (37), for the first time in 2007, showed that miR-200c overexpression led to decreased ZEB1 gene expression and increased E-cadherin expression in BC cell lines. Also, many reports have shown that miR-200c has EMT-dependent tumor suppressor function in BC (38, 39). The miR-200c may have a protective effect against BC, leading to an increase in the patients’ survival time. Marchini et al. suggested that patients with high levels of miR-200c expression had a better prognosis and longer survival than patients with low expression levels in stage I ovarian cancer (40). Furthermore, the miR-200c expression showed a significant decrease in colon cancer. Its expression is also inversely related to tumor size, serous attack, lymph node metastasis, and tumor node metastasis (41).
The results of the present study are in line with previous studies demonstrating that the miR-200c expression as a tumor suppressor is decreased in BC compared to normal tissues; thus, these microRNAs play an important role in metastasis and invasive BCs.
In conclusion, we examined miR-30a and miR-200c expression levels and realized that miR-30a and miR-200c in stages III and IV had lower expression levels than stages I and II. The results of this study also showed that the expression levels of miR-30a and miR-200c were associated with tumor metastasis to lymphatic vessels in patients. In this regard, these two miRNAs showed decreased expression levels in patients with lymph node metastasis compared to patients with no lymph node metastasis, indicating a strong association between these biomarkers and BC. These results suggest that these biomarkers can be specifically used to screen for BC in different stages. In general, identifying the association between miRNA expression levels and clinical-pathological features of BC patients is important and may lead to the introduction of a diagnostic biomarker.