1. Context
Among regenerative capabilities, the ability to heal spinal cord injury (SCI) is an ability that is lost in most vertebrates, especially mammals. In addition, studying this process can yield valuable knowledge for human beings to regain this lost ability. Currently, the approach to an injured spinal cord is securing a vulnerable cord and controlling vital signs. Surgical decompression and rehabilitation is still the most reliable treatment approach in SCI for humans. On the other hand, there are species other than human beings from the Animalia kingdom capable of regenerating their spinal cord after injuries, such as planarians, vertebrate zebrafish, and Xenopus laevis (1-3). Some of these animals can regenerate their injured spinal cord at any time of their life. Planarians, for example, benefit from their stem cells, called neoblasts, which can replace damaged cells. Their regenerative capacity is significant in that they can regenerate most of their nervous system after injury, including the whole brain (3).
It should be noted that the planarians are invertebrates and have two ventral nerve cords with multiple transverse nerves and an anterior brain (4). However, investigating molecular pathways and genes involved in its nerve cord repair can yield significant clues. In addition, the teleost fishes, including the zebrafish, are among the vertebrates that can gain their function after SCI. The zebrafish is accepted as a model of spinal cord regeneration worldwide due to its ability to regrow and remyelinate axons (5).
On the other hand, there are some species that benefit from their regenerative capabilities for a while but lose their capability in another period of life. Xenopus laevis is among these animals. The tadpole (immature animal) is in the regenerative phase: It is capable of regenerating a functional spinal cord after transection or injury. However, the froglet (mature animal) is in the non-regenerative phase and loses the ability to recover after SCI, and the animal has a refractory period, which can only regenerate its tail without neural regeneration. Studies have shown that the regeneration can be reactivated in this period by activating BMP or Notch signaling pathways (6).
Some animals have spinal cord regenerative capability for all of their life; others have limited spinal cord regenerative capability, and others have no regenerative capability at all. Studying the genes and pathways involved in SCI regeneration in regenerative animals and their alterations during metamorphosis and the loss of SCI recovery ability will give better insight into the loss of regeneration ability through evolution (2).
Among all the species, zebrafish is an animal widely studied due to its SCI regeneration capacity and being known as an animal model for SCI regeneration. Studying the pathways that lead to SCI recovery and the alterations leading to the loss of this ability can also guide us in extrapolating this capability to human beings. The aim of this study is to review the gene-based studies in regenerative animals, including zebrafish, X. laevis, and planarians, and compare their expression patterns and fold-change slopes to the non-regenerative ones to determine key points in the process of SCI recovery. Possible opportunities to translate the current knowledge to the human species are also discussed.
2. Evidence Acquisition
This review has been performed according to the PRISMA checklist (7). To include all the eligible published studies on the gene expression of X. laevis, zebrafish, and planarians during the SCI, a systematic search was performed with the assistance of an expert librarian. Medline, Embase, Scopus, CENTRAL, PROSPERO, and JBI Database of the Systematic Reviews with the search query of “(Spinal cord injury*) AND (Gene expression) AND ((Xenopus laevis) OR Planarian OR zebrafish OR Zebra fish)”. The references of review articles were also searched for higher sensitivity.
Data collection was performed using Aromataris’ suggested method (8). Concisely, the studies were transferred to the 2019 edition of the Endnote software. The researchers utilized the software’s feature to eliminate duplicate studies. Subsequently, the studies were evaluated by two separate researchers based on their titles and abstracts. The discrepancies were settled by the intervention of a neutral third party. The data were extracted to a predesigned Microsoft Excel Worksheet with an emphasis on the authors and their funding (if any), publication year, study design, sample size, studied species, studied genes, nature of the injury and its level, and gene expression alterations after the injury.
The inclusion criteria were as follows:
- The study should be on the mature and immature X. laevis, planarian, or zebrafish with SCI.
- The study should evaluate a gene expression pattern and compare it to a control group or the pre-injury state.
- The process assessment of all the studies that were included was conducted using appropriate laboratory techniques, including polymerase chain reaction (PCR) or real-time PCR, immunohistochemistry, western blot, immunostaining, in-situ hybridization, and protein quantification.
The exclusion criteria were studies with pharmacological or other types of intervention on the above-mentioned animals, and any studies conducted without appropriate laboratory methodologies and without comparison to a control group or pre-injury state were eliminated.
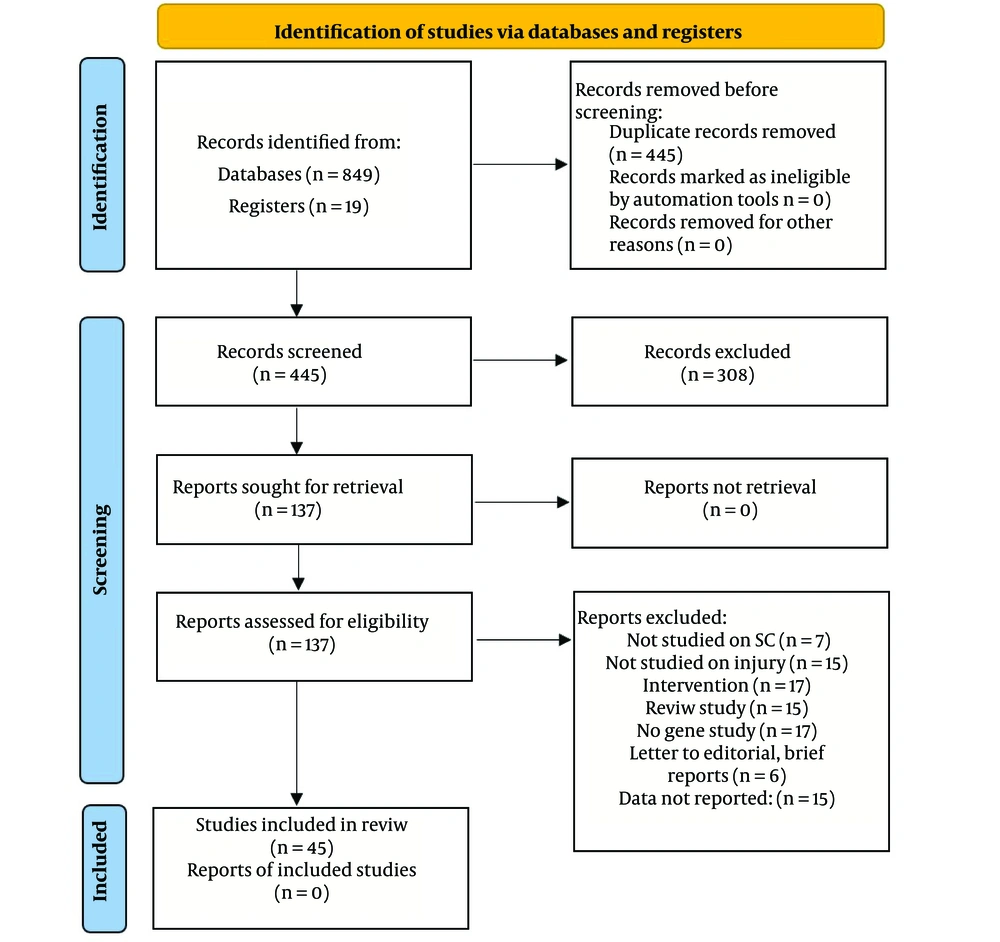
As a result of this process, 45 studies were included in the extraction step (Figure 1). The homologous/paralogous genes were found in the National Center for Biotechnology Information (NCBI) Gene and the Ensembl databases.
Two major genome-wide studies were also included in the paper (1, 2), one for the zebrafish and one for the X. laevis, and the authors decided to pool these data separately. After initial processing, 238 common genes were detected in the studies. The zebrafish study reported gene expression in post-injury days 1, 3, and 7, and the X. laevis study reported them in post-injury days 1, 2, and 6. The dates are labeled as early phase, intermediate phase, and late phase, respectively. The expressions were then labeled as “up-regulated" or “down-regulated" in the early, intermediate, and late phases. The similarity of these regulations (similar up- or down-regulation in each time phase) in zebrafish or regenerative X. laevis and their contrast with non-regenerative X. laevis were marked as “significant patterns." A total of 9 significant patterns were found among the genes (similarity of the zebrafish and the regenerative X. laevis in early, intermediate, or late phase [3 significant patterns], and contrast between the early, intermediate, and late phases of the non-regenerative with regenerative X. laevis or the zebrafish [6 significant patterns]).
The same process was performed for the slope of the expression patterns. The expression of each gene can elevate or decline between two phases, regardless of its original level. An expression fold-change from 1.5 to 2 from the early to intermediate phase or the intermediate to late phase was considered an ascending slope, as was the change from -1 to -0.5 of -1 to +2. A similar early-to-intermediate or intermediate-to-late phase slope between the zebrafish and the regenerative X. laevis was considered a significant slope pattern. On the other hand, the contrast of the slope between the zebrafish or regenerative X. laevis with the non-regenerative X. was also labeled as other significant slope patterns, thereby yielding 6 significant slope patterns.
3. Results and Discussion
The capability or incapability of regenerating the spinal cord has been studied in a variety of animals, including the newt, axolotl, lamprey, mouse, X. laevis, and zebrafish (9-12). However, this paper has focused on zebrafish and regenerative and non-regenerative X. laevis. However, there are other studies focusing on the inhibitory genes, such as the Rab27b gene, reported by Sekine et al., whose ablation can enhance axonal regeneration in mice tissue (12). Overall, 45 studies were included for full-text analysis: 1 study on the planarian, 36 studies on the zebrafish, and 8 studies on the regenerative and non-regenerative X. laevis response to SCI. A summary of the study characteristics and their brief findings are depicted in Figure 1. Among these studies, 112 genes were extracted and identified as significant genes in the process of SCI recovery, summarized in the supplementary file, and the characteristics of the included studies are shown in Table 1.
| N | Study | Species | Gene(s) (NIH Gene ID) | Follow-up(s) | Site of Injury | Measurement Method | General Finding(s) |
|---|---|---|---|---|---|---|---|
| 1 | Barreiro-Iglesias et al. 2015, (13) | Mature zebrafish | 5-HT1A (100134982) | 14 days | 3.5 mm caudal to the brainstem-spinal cord junction | Immunohistochemistry and real-time PCR | Serotonin, released by serotonergic neurons, induces motor neuron regeneration after spinal cord injury. |
| 2 | Becker et al. 1998, (14) | Mature zebrafish | L1.1, L1.2, and NCAM (30656, 30634, 30447) | 7 and 14 days | 3.5 mm caudal to the brainstem/spinal cord transition zone | In-situ hybridization | The L1.1 gene was weakly upregulated 14 days after injury. The L1.2 gene was, however, upregulated at both times. There was no difference in the NCAM gene. |
| 3 | Becker et al. 2005, (15) | Mature zebrafish | GAP-43 and L1.1 (30608, 30656) | 6 and 18 days | Operculum and 4 mm and 8 mm caudal to operculum | Real-time PCR | Both genes were upregulated. GAP-43 expression did not change over time. However, its overexpression was lower in lower injuries. On the other hand, the L1.1 gene reduces from the 6th to the 18th day. |
| 4 | Briona et al. 2015, (16) | Mature zebrafish | Wnt signaling | 1, 3, 5, and 7 days | The level of the anal pore | Immunohistochemistry | Wnt reporter is a necessary element for SC regeneration from the 1st to the 7th days after injury. The signaling mediators are dominantly expressed by the glial cells. |
| 5 | Cebria et al. 2002, (4) | Planarian | 953-HH, 721-HH, 1791-HH, 517-HH, 2467-HH, 1242-HH, 1008-HH, 944-HH, Eye793, 5189-HH, Eye53, and 1020-HH gene | 1 to 5 days | Parapharyngeal level | Immunostaining | A three-stage regeneration model is suggested as the appearance of new brain primordium in a regenerative blastemal, differentiation, and structural restoration. |
| 6 | Chen et al. 2016, (17) | Mature zebrafish | L1.2 (30634) | 4 and 12 hours, and 6 and 11 days | 4 mm caudal to the brainstem-spinal cord transitional junction | Real-time PCR and in-situ hybridization | The L1.2 gene did not change in the first hours but upregulated in the following weeks in comparison to sham group. |
| 7 | Dias et al. 2012, (18) | Adult zebrafish | Her 4.1 and notch1b (100149863, 794892) | Up to 14 days | 4 mm caudal to the brainstem-spinal cord transitional junction | In-situ hybridization and immunohistochemistry | Notch acts as a negative factor for the proliferation of the progenitor cells and the motor neuron generation. |
| 8 | Dupret et al. 2017, (19) | Immature zebrafish | Ezh2 (768133) | 6 and 9 days | Within the pigment gap distal to the circulating blood | Real-time PCR | The gene was expressed in the regenerating caudal fin and its spinal cord after amputation. |
| 9 | Gaete et al. 2012, (20) | Xenopus tadpole | Sox2 (398000) | 6 and 8 days after amputation | Middle of the tail | Real-time PCR, in-situ hybridization, western blot, and immunohistochemistry | Sox2 and Sox2+ cells are required for SC regeneration and amplify immediately after the SCI. |
| 10 | Goldshmit et al. 2012, (21) | Mature zebrafish | LPA1 (368461) | Not mentioned | Between the dorsal fin and the operculum, corresponding to the eighth vertebra (5 mm caudal to the operculum) of the spinal cord | Real-time PCR and in-situ hybridization | The gene was upregulated after the injury. |
| 11 | Goldshmit et al. 2018, (22) | Mature zebrafish | fgf8a, fgf3, pea3, erm, spryd4, FgfR2, p-MAPK, and Nestin (30538, 30549, 30700, 30452, 100005939, 352940, 65237, 100150939) | 3 days, 2 and 3 weeks | Within the eighth vertebra | Real-time PCR | The genes are part of the fgf signaling pathway. |
| 12 | Guo et al. 2011, (23) | Mature zebrafish | Sox11b, Ascl1a, and Nestin (30603, 30466, 100150939 | 4 and 12 hours, and 11 days | 4 mm caudal to brainstem/spinal zone | Real-time PCR | Asc11a was upregulated to 2 folds in all of the time points. However, the Sox11b and Nestin genes were upregulated significantly only on the 11th day. |
| 13 | Hatta-Kobayashi et al. 2016, (24) | Refractory and non-refractory Xenopus laevis | Xenopus neuronal pentraxin I (xNP1) (108697994) | 10, 24, and 48 hours | 2-mm wide tail tissues from the end of the amputated tail stumps | In-situ hybridization | The gene was downregulated in the refractory state but upregulated in the non-refractory phase. |
| 14 | Hui et al. 2014, (1) | Mature zebrafish | Hsp90a1, hsp90a2 (30591, 565155) | 1 day | Within 15th-16th vertebra | Real-time PCR | The genes were upregulated on the first day but downregulated on the days after that. |
| 15 | Kuscha et al. 2012, (25) | Mature zebrafish | Vsx1 (30598) | 2 weeks | 3.5 mm caudal to the brainstem-spinal cord junction | Western blot | Vsx1 gene was upregulated 2 weeks after injury. |
| 16 | Lee-Liu et al. 2014, (2) | Mature and immature Xenopus laevis | This study was a “genome-wide expression" profiling | 1, 2, and 6 days. | At the midpoint between the fore and hind limbs (mid-thoracic, approximately between the seventh and eighth vertebrae) in NR-stage and limb buds were used as a reference in the R-stage | Real-time PCR | R-stage showed different patterns of gene expression in both time and transcripts from the NR-stage. |
| 17 | Lin et al. 2012, (26) | Adult zebrafish | Contactin-2 (30726) | 4 and 12 hours, and 6 and 11 days | 4 mm caudal to the brainstem spinal cord transitional junction | Real-time PCR, in-situ hybridization, western blot, and immunohistochemistry | Contractin-2 upregulates 6 and 11 days after injury, which has similar localization with the motor neurons. |
| 18 | Liu et al. 2016, (27) | Adult zebrafish | Melanoma cell adhesion molecule (338313) | 4 and 12 hours, and 6 and 11 days | 4 mm caudal to the brainstem spinal cord transitional junction | Real-time PCR, in-situ hybridization, western blot, and immunohistochemistry | The MCAM mRNA was upregulated from the beginning of the injury and was significantly higher than the sham group in the 6th and 11th days. |
| 19 | Liu et al. 2014, (28) | Mature zebrafish | Ptena and Ptenb (794088, 368415) | 12 hours and 6 days | 4 mm caudal to the brainstem spinal cord transitional junction | Western blot | The Ptena gene was upregulated in both time points. Ptenb was also upregulated during the first 12 hours after injury. |
| 20 | Ma et al. 2012, (29) | Mature zebrafish | Csrp1a (378726) | 4 and 12 hours, 3, 11, 21 days | Through the second and the eighth vertebrae, | Protein quantification | The gene was upregulated in descending axons in the 3rd, 11th, and 21st days. |
| 21 | Ma et al. 2014, (30) | Mature zebrafish | Legumain (406625) | 4 and 12 hours, and 11 days | Between the eighth and the ninth vertebra | Western blot | There was no significant difference in the gene expression in the early phase. However, it was significantly upregulated on the 11th day. |
| 22 | Ma et al. 2016, (31) | Mature zebrafish | Glucuronyl transferase (GlcAT-P) and human natural killer cell antigen-1 sulfotransferase (HNK-1ST) (548607, 445322) | 1, 3, and 11 days | Between the eighth and ninth vertebrae, 3.5 mm caudal to the brainstem-spinal cord junction | In-situ hybridization | The genes were both upregulated on the 11th day but not the 1st or 3rd day. |
| 23 | Mokalled et al. 2016, (32) | Mature zebrafish | Fibronectin 1a (fn1a), fibronectin 1b (fn1b), connective tissue growth factor a (ctgfa), myostatin b (mstnb), stanniocalcin 1 like (stc1l), and GFAP (100005469, 334613, 321449, 798441, 393511, 30646) | 1, 2, and 5 weeks | Not mentioned | In-situ hybridization and immunohistochemistry | The so-called genes were upregulated in the follow-up times. |
| 24 | Munoz et al. 2015, (33) | Mature and immature Xenopus laevis | Sox2 and Sox3 (398000, 399335) | 1, 2, or 6 days | Mid-thoracic level | Real-time PCR | The gene expression did not change significantly during the metamorphisms. |
| 25 | Ogai et al. 2014, (34) | Adult zebrafish | Sox2 (378723) | 3, 5, 10, 15, and 20 days | 4 mm caudal to the brainstem spinal cord transitional junction | Real-time PCR and immunohistochemistry | The Sox2 was significantly upregulated just after the injury. |
| 26 | Pan et al. 2013, (35) | Mature zebrafish | MVP, Islet-1, TH, and Nestin (373081, 30147, 30384, 100150939) | 4 and 12 hours, and 6 and 11 days | 4 mm caudal to the brainstem-spinal cord transitional junction | Real-time PCR, in-situ hybridization, western blot, and immunohistochemistry | The MVP was upregulated, not in the first hours but through the weeks. (Western blot showed upregulation, even in the first hours.). Islet-1, TH, and Nestin were also upregulated in the 11th day. |
| 27 | Peng et al. 2017, (36) | Adult zebrafish | Sema4D (100333323) | 4 and 12 hours, and 3,11, and 21 days | 4 mm caudal to the brainstem spinal cord transitional junction | Real-time PCR, in-situ hybridization, and immunohistochemistry | The gene was upregulated in the acute response phase (within 3dpi) and downregulated in the chronic response phase (11–21dpi) |
| 28 | Reimer et al. 2009, (37) | Mature zebrafish | Olig2, shha, smoothened, patched1, fgf3, spry4, rarab, rxrga, rxrgb, crapb2a, cyp26a (325288, 30269, 30225, 30189, 30549, 114437, 555364, 30464, 436617, 324340) | 2 weeks | 4 mm caudal to the brainstem-spinal cord transitional junction | Real-time PCR and in-situ hybridization | All the so-called genes were upregulated 2 weeks after the injury. |
| 29 | Reimer et al. 2013, (38) | Mature zebrafish | D4a and patched2 (30329, 30181) | 14 days | 4 mm caudal to the junction between brain stem and spinal cord | Real-time PCR | The genes upregulated rostral but not caudal to the lesion site. |
| 30 | Ribeiro et al. 2017, (39) | Adult zebrafish | Foxj1a (767737) | 3, 7, 14, and 30 days | The spinal cord was compressed dorsoventrally using forceps | In-situ hybridization and immunohistochemistry | Foxj1a is required for ependymal cell positioning. It is also actively associated with ependymal cell proliferation during SC regeneration. |
| 31 | Sasagawa et al. 2016, (40) | Immature zebrafish | E2F4, TFDP1, FOXM1 (406741, 393749, 394072) | 1 and 3 days after injury | Above the caudal end of the swim bladder. | In-situ hybridization | The genes possibly regulate zebrafish SCI-specific differentially expressed genes. |
| 32 | Schweitzer et al. 2007, (41) | Adult zebrafish | Contactin1a (353150) | Up to 180 days | 8th vertebrae, between the dorsal fin and the operculum | Real-time PCR, in-situ hybridization, and immunohistochemistry | The gene was upregulated at the 6th and 14th days. |
| 33 | Strand, 2016, (42) | Mature zebrafish | Wnt4b (791993) | 3 and 14 days | At the level of the 8th vertebra | Real-time PCR | The gene expression was increased in the injured fishes in comparison with the sham group. |
| 34 | Strand et al. 2016, (43) | Adult zebrafish | Wnt/beta-catenin signaling | 3, 14, and 21 days after injury | 8th vertebrae, between the dorsal fin and the operculum | Immunohistochemistry and real-time PCR | The Wnt signaling was increased from the first 3 days and was significantly higher than the sham group after 14 days. |
| 35 | Tapia et al. 2017, (44) | Mature Xenopus laevis | Lif, leptin, and socs3 (100381055, 108711253, 414533) | 3, 6, 24 hours, 2, 6, 15, 30 days | Midpoint between forelimbs and hindlimbs | Western blot | The genes are of the JAK-STAT pathway. The difference between the R-stage and NR-stage was not the time but the phenotype of pSTAT3+ motoneurons. |
| 36 | Wang et al. 2017, (45) | Mature zebrafish | ATF3 (393939) | 12 hours, and 6 and 11 days | At the level of the 8th vertebra | Western blot | The gene was upregulated in the first 12 hours and the 6th day. However, it was normalized until the 11th day. |
| 37 | Wehner et al. 2017, (46) | Mature and immature zebrafish | Mature: Collagen1a2 (col1a2) and fibronectin1a (fn1a) Immature: Wnt/β-catenin (336471, 100005469) | Mature: 2 days Immature: 1, 2, 3, and 5 days | Mature: 3.5 mm caudal to the brainstem-spinal cord junction; Immature: Anal pore immediately dorsal to the notochord | In-situ hybridization | All the so-called genes were upregulated in every stage. However, the Wnt/β-catenin gene was normalized 5 days after the lesion. |
| 38 | Yu et al. 2011, (47) | Mature zebrafish | Tenascin-C (30037) | 4, 12, 24, and 48 hours, and 11 days | At the level of the 4th vertebra | Western blot | The gene expression was elevated in the first 4 hours, which was decreased until the 24th hour. It was secondly elevated in the 48th hour and was high until the 11th day. |
| 39 | Yu and Schachner, 2013, (48) | Mature zebrafish | Syntenin-a (325004) | 4 and 12 hours, and 6 and 11 days | At the level of the 4th vertebra | Western blot | The gene was upregulated in all of the time points. |
| 40 | Shao et al. 2023, (49) | Zebrafish | DUSP2 (445057) | 8 days | Spinal cord | Western blot, qPCR | DUSP2 up-regulates after spinal cord injury. DUSP2 retards axonal regeneration through JNK phosphorylation. |
| 41 | Hosseini et al. 2022, (50) | Zebrafish | drd2a (282557), drd2b (378719), drd3 (282554), drd4a (503564), drd4b (503565) | 1, 7, and 14 days | 9th - 10th vertebra | qPCR | The dopaminergic receptors upregulated 7 days after injury. drd2a and drd2b were mostly upregulated in females |
| 42 | Torruella-Gonzalez et al. 2022, (51) | Xenopus laevis | Cornifelin (444756) | 6 days and 10 days | Spinal cord | qPCR | Cornifelin was detected in the meningeal tissue 6 days after injury. The gene RNA was also detected in the injured site 10 days after injury. Cornifelin was detected weaker in adult Xenopus laevis. |
| 43 | Pelzer et al. 2021, (52) | Immature Xenopus laevis | Foxm1 (496385) | 0, 1, and 3 days | Spinal cord | qPCR | FOXM1 was upregulated 3 days after injury. FOXM1-knockout tadpoles had weaker recovery. |
| 44 | Cui et al. 2021, (53) | Zebrafish | NPY1R (793668) | 12 hours, 6 days, and 21 days | Spinal cord | qPCR, in-situ hybridization, and immunostaining | Neuropeptide Y is decreased, and its receptors are down-regulated in early and intermediate phases post-injury. |
| 45 | Cavone et al. 2021, (54) | Zebrafish | TNF Signaling pathway | 1 and 3 days | Spinal cord | qPCR | The TNF-α pathway products secreted by the macrophages promote progenitor cells. TNF promotes hdac1 production in progenitor cells to promote neurogenesis. |
A Summary of the Included Studies and Their Characteristics
Two studies were genome-wide studies about the zebrafish and the X. laevis that were compared separately (1, 2).
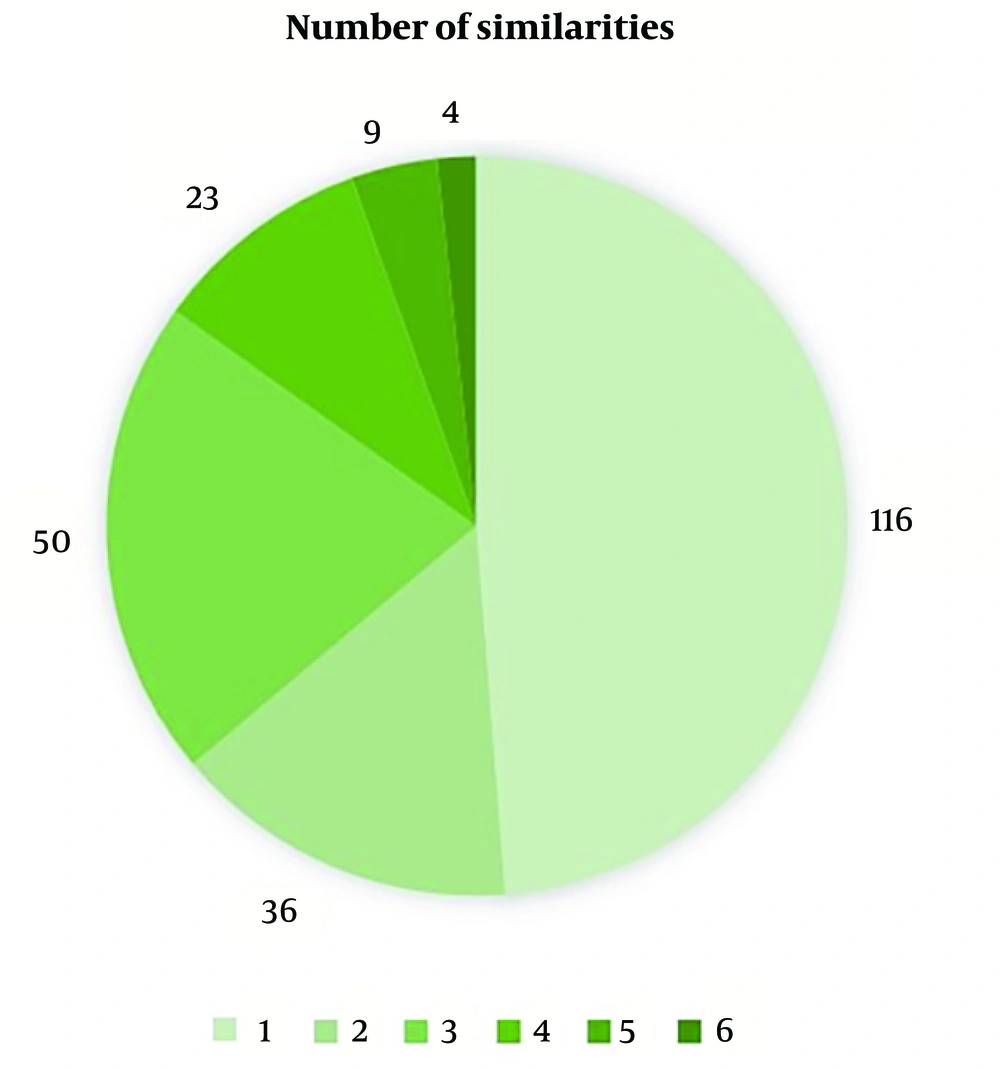
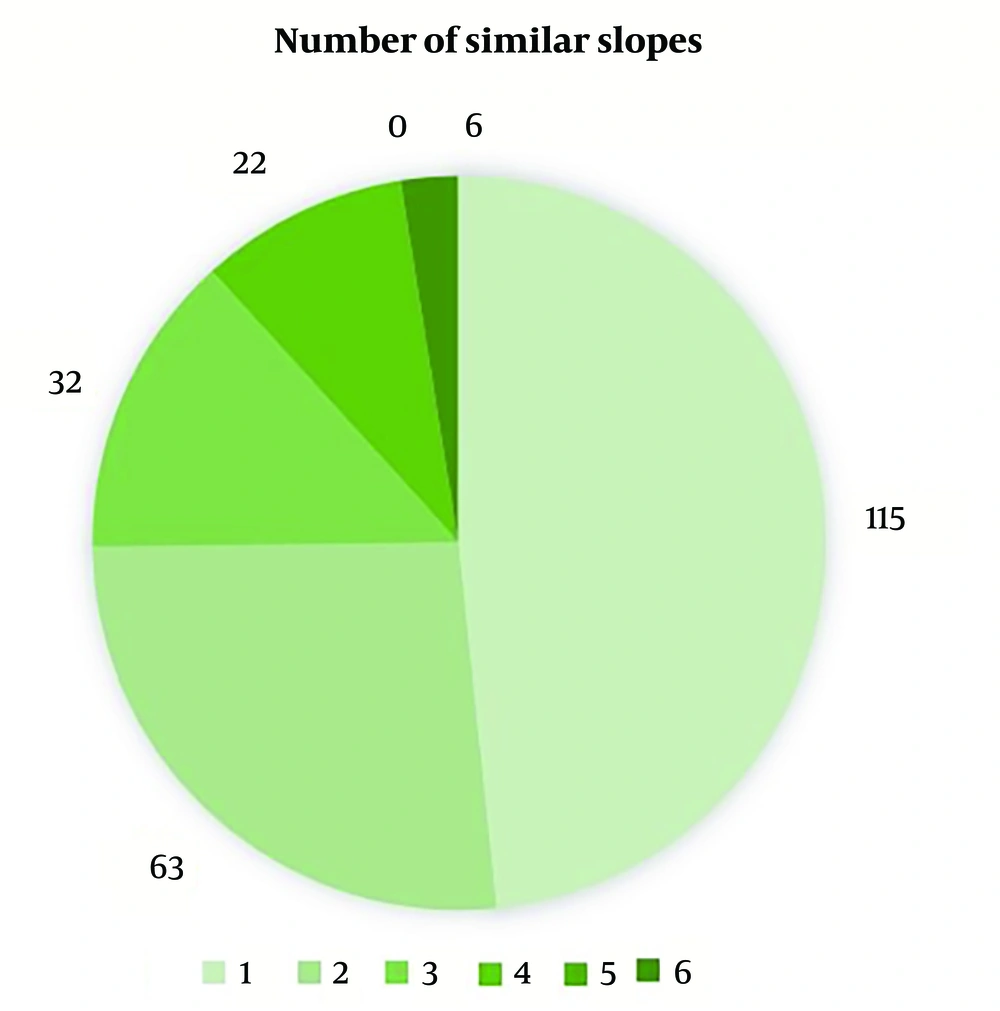
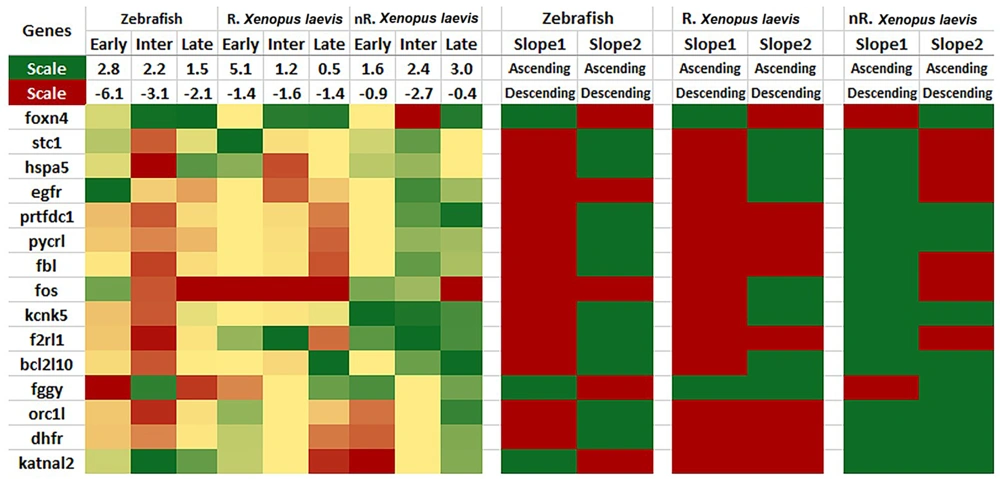
After initial processing, 238 genes were common among these studies. As mentioned in the Methods section, 9 significant expression patterns were possible (indicating similar directions in the regenerative animals and opposing directions in the non-regenerative ones). Among these 238 common genes, genes 23, 9, and 4 followed 4, 5, and 6 of these significant patterns, respectively (Figure 2). Similarly, 6 of these genes followed all six of the significant slope patterns, none followed five of them, and 22 followed four of them (Figure 3). After gene-by-gene investigations, there were 15 genes with more than 4 significant patterns, both in expression and slope (more than 8 significant patterns as a whole). The expression patterns of these 15 top genes are also shown in Figure 4 (based on the data reported in the original papers), and the full list of the genes with their comparisons is shown in Appendix 1. According to these data, FOXN4, STC1, HSPA5, EGFR, PRTFDC1, PYCRL, FBL, FOS, KCNK5, F2RL1, BCL2L10, FGGY, ORC1L, DHFR, and KATNAL2 genes have a significantly similar expression pattern in the regenerative animals and a significantly opposing pattern in the non-regenerative ones.
Heat map of the top 15 genes. On the left side, the foldchange of the genes in each phase of the species is shown in green (up-regulation), red (down-regulation), or yellow (neutral). The scale of each column is located in the top row. On the right, the ascending (green) or descending (red) slope of each gene is demonstrated for each species (slope 1: Early-to-intermediate phase slope; Slope 2: Intermediate-to-late phase slope; R: Regenerative; nR: Non-regenerative.)
3.1. Top Common Genes Between the Zebrafish and Xenopus laevis
Regarding these two genome-wide studies, there were 238 common genes among the zebrafish and X. laevis, and there were not any common genes between these two studies and the other 37 studies. A total of 238 common genes were analyzed to determine which show the same expression pattern in immature X. laevis and zebrafish and a different expression pattern in mature X. laevis. Two significant patterns were considered in the analysis, one with the same expression change in the three time-points (Appendix 1 and Figure 2) and the other with the slope of expression between the three time-points (Figure 3). Fifteen top common genes were found between the two significant patterns. These genes are shown as a heat map in Figure 4.
The top gene in this analysis, FOXN4, is a member of the forkhead/winged-helix family, which has an important role in cell differentiation, developmental processes, and neurogenesis in the central nervous system (CNS). Its expression pattern in different phases shows upregulation in regenerative animals (zebrafish and immature X. laevis) and downregulation in nonregenerative animals in the intermediate phase (52, 55, 56). N4 transcription was also studied after SCI in adult rats, and its expression pattern was similar to zebrafish and immature X. laevis, implying the promotion of spinal cord regeneration in mammals. This gene participates in astrocyte proliferation after SCI in adult rats (57).
The STC1 gene is also one of the top 10 differentially regulated transcripts one day post-injury in X. laevis, in that its fold change ratio (FC between regenerative and nonregenerative, FC(R)/FC(NR)) is reported as 27.06. This gene shows upregulation in the intermediate phase in nonregenerative X. laevis but downregulation in zebrafish. The STC1 gene also demonstrates a negative slope between the early and intermediate phases in both regenerative animals and a positive slope in nonregenerative X. laevis.
The HSPA5 gene is categorized within the group of “response to stress" genes and was upregulated in the early phase in all three animals, with the greatest upregulation occurring in the regenerative stage of X. laevis. The expression pattern shows a negative slope between the early and intermediate phases in regenerative animals but a positive slope in nonregenerative X. laevis. The HSPA5 gene has been evaluated in an in vitro rat model of SCI and is upregulated one day after injury, peaking at 4 hours post-injury. This gene is involved in the PI3K/AKT/mTOR signaling pathway of endoplasmic reticulum in response to stress and promotes autophagy after injury. Altogether, HSPA5 is upregulated in both regenerative animals in the early stage after mechanical injury and has a positive effect on regeneration via neuronal protection (58).
In addition to the genes upregulated during the regeneration phase, the pattern of gene expression in non-regenerative X. laevis should also be mentioned. For example, the Fos proto-oncogene (FOS gene) can dimerize with proteins of the JUN family, forming the AP-1 transcription factor. As a result, the FOS proteins are known as regulators of cell proliferation, differentiation, and apoptosis (54). As shown in Figure 4, this gene down-regulates and then up-regulates during early and late phases in regenerative X. laevis, respectively. However, the genes show a totally contrasting pattern in non-regenerative X. laevis with early up-regulation and late down-regulation. The epidermal growth factor receptor (EGFR) gene is also another gene with significant up-regulation in non-regenerative X. laevis but down-regulation in zebrafish and regenerative X. laevis. Moderating these genes and their activity can be key in promoting the regeneration of an injured spinal cord.
3.2. The Role of Identified Genes in the Process of Spinal Cord Regeneration
Based on the findings of the included studies, it is suggested that SCI regeneration involves cell proliferation, differentiation, alignment, and plasticity, all of which are modulated by the cellular microenvironment. Monitoring and analyzing the process of healing, from the first moment of injury to the last day of regeneration, brings the idea of considering the microenvironment a timeline (4). This microenvironment is mostly mediated by glial or fibroblast-like cells during cell proliferation, differentiation, and alignment (32, 35, 38, 43). During the proliferation step, there are many different cytokines, growth factors, and tumor suppressors that play a role in the early response to SCI. E2F transcription factors and related genes, such as TFDP1, FOXM1, and FOXN4, are among the genes expressed within the first days after injury in zebrafish (1, 40). These genes are believed to play a role in the progression of cells from the G0 to M and S1 phases of the cell cycle (59, 60). The CHAF1B gene, another cell-cycle modulator, was also identified in the genome-wide study of X. laevis as being overexpressed in the first days after injury. However, the gene was down-regulated at the same time among the animals in the non-regenerative state(2). Controlling cell life is another aspect of this initial step. SOX and JAK-STAT-related products, such as leptin, LIF, of SOCS3, were important in the early response to SCI. The opposite pattern was reported in non-regenerative X. laevis (33, 44).
Differentiation is another important step in the process of SCI recovery. Some of the role-players in this process are ASCL1 and Crapb2a genes and the fibroblast growth factor signaling genes FGF3, FGF8, FGF17, FGFr1, FGFr2, or FGFr4 (22, 23, 37, 61). Retinoic acid and its metabolites are known as an inducer of neuronal development (61).
Most of the genes related to retinoic acid metabolism were among the significant genes in the early and intermediate phases of the SCI recovery, including Crapb2, Raldh2, Raraa, Rarab, Rarga, Rargb, Rxraa, Rxrga, and Rxrgb (37). Rbp4 was also one of the common genes found in the genome-wide studies regulating retinoic acid metabolism. Rbp4 is down-regulated in all of the first 7 post-injury days in the zebrafish and the regenerative X. laevis. In contrast, the gene was significantly up-regulated in the same days in the non-regenerative X. laevis (1, 2).
After the proliferation and differentiation phases, nascent cells must be maintained for correct alignment and branching. This step, considered an intermediate reaction rather than an early one, is also orchestrated by many genes identified in this review. Dopamine and its related genes, such as MVP and DRD4, as well as tyrosine hydroxylase and serotonin, are found to be important in the alignment of the new cells (13, 35, 38, 50). Dopamine, for example, is secreted mostly proximal to the injury, which leads to neurogenesis. Blocking or distorting the balance of dopaminergic receptors yields insufficient motor recovery in the zebrafish (38). In addition to alignment, proteins such as Csrp1, Tenascin C, or SDCBP induce plasticity and the creation of new synapses during this step (29, 47, 48). The aforementioned genes are over-expressed on the third-day post-injury and are down-regulated after 2 weeks. After this step, anti-inflammatory cytokines will be overexpressed, and promoting factors will be suppressed in order to regain full function after SCI (1, 15, 31).
The microenvironment of the injury site is a key factor in these processes. The secretion of angiogenic factors, such as MCAM, by fibroblasts and glial cells at the injury site in the intermediate and late phases, in addition to the elevation of proliferating agents in the early phase and their drop in the late phase, enriches the microenvironment with collagen filaments and fortifies it with important factors such as retinoic acid metabolites (27, 32, 46). These glial-derived factors are also among the main differences between the regenerative and non-regenerative X. laevis (2).
A considerable number of the genes reviewed in the supplementary file are expressed in non-neuronal cells. The connective tissue growth factor Erm, the FGF family, and GFAP are mostly secreted by glial cells in the CNS (32, 62). The Dkkb gene, responsible for locomotor recovery and glial bridge formation, is also present in fibroblast-like cells (16). The fibronectin family genes, associated with late recovery and motor function improvement, are expressed in ependymal cells, and the Fox genes, upregulated in the early recovery, are responsible for ependymal cell expansion(32, 39). Oligodendrocytes and Schwann cells also play an early role in eliminating dead cells through lysophosphatidic acid (LPA) signaling (21). On the other hand, the immune cells are also other types of cells that affect the microenvironment not only by affecting the fibroblast's activity (which is discussed before) but also by their own actions as the HNK-1ST gene shown to upregulate in the late-phase of regeneration in the zebrafish (31). These findings highlight the importance of non-neuron cells in the CNS and their contribution to neuronal regeneration.
Healing an injured spinal cord is a multifactorial process that some animals have retained, and others have lost through evolution. This study reviewed the current literature on genes related to spinal cord regeneration and their expression patterns from the initial injury to the last recovery day. Heterogeneity is one of the greatest concerns in this study. As we have limited data to follow spinal cord regeneration through the phylogenetic tree, the authors have selected three points in the hope of finding light through the way of genetic investigation of the spinal cord regeneration process. Further biological and bioinformatics studies are needed to confirm the homology of the identified genes and their value in the newer species. The applicability of this knowledge to other animals and to the human being should be tested.
Gene engineering techniques or miRNA attenuation can be tested to stimulate regenerative potential in other species. Some genes, such as HSPA5, c-FOS, and the BCL family (as contributors to the process of apoptosis), should also be studied more in-depth for this purpose. It should be considered that this study has only evaluated and compared the gene expression patterns, and the functional role of the specified genes might vary. Experimental in-vitro or in-silico studies are needed to confirm the validity of the role of these genes in the process of spinal cord regeneration. Most of the studies only reported comparisons, and full expression patterns or fold-change data of each day were not reported. However, a list of promising genes and pathways are presented in this paper that can be further studied in the future to advance the healing of an injured spinal cord in non-regenerative species.
4. Conclusions
As demonstrated in the supplementary file, most relevant genes detected in these studies have homologous or orthologous genes in human beings. However, in some cases, their expression pattern differs.
CCN2, an important gene for the cellular proliferation and creation of the glial bridge, has significantly lower expression in the human CNS (32, 63). Wnt and E2F signaling-related genes, such as DKK1, FOXN4, FOXM1, and TFDP1, important cell cycle mediators that lead cells from G0 to M and S1 phases, are also active in human beings. However, these genes are mainly expressed in the placenta, testis, and bone marrow, with insignificant activity in the CNS (40, 43, 52, 63). The EGFR, c-FOS, and ORC11 genes should also be mentioned as important cell-cycle modulators, according to genome-wide studies (1, 2).
The timing and creation of an appropriate microenvironment by genes, such as MCAM and Dkkb, is critical to SCI healing in the regenerative X. laevis and the zebrafish. Nevertheless, an inadequate microenvironment is a common factor between human beings and the non-regenerative X. laevis (1, 2, 44, 51). In the early phase, controlling inflammation and scar formation, as well as creating a rich environment for progenitor cell proliferation and differentiation, are important for neuronal alignment, branching, and synapse formation in the intermediate phase. Key activities of the regenerative animals also include suppressing early-phase agents and enhancing the neuroplasticity of the injury site in the late phase, which is not seen in non-regenerative models of SCI.
The additional detection of potential candidates for each step of SCI healing, in addition to timing or altering the expression of important genes and establishing an appropriate microenvironment through genetic engineering, is paramount to optimizing SCI recovery in human beings.