1. Introduction
About 1500 species of infectious organisms are known to be pathogenic to humans, including 217 viruses and prions (1, 2). Unpredictable hemorrhagic fever disease outbreaks are caused by different species of Ebola viruses, as well as Marburg virus associated with 90% fatality rates (3). The ongoing multinational Ebola outbreak posed a serious threat to the countries of West Africa and beyond due to its unprecedented magnitude. Ebola is an enveloped virus that belongs to the family Filoviridae. It includes five species of Ebola and a strain of Marburg virus, which cause hemorrhagic fever in humans and also in nonhuman primates. The term viral hemorrhagic fever refers to a variety of viral diseases characterized by bleeding and fever in humans. This syndrome is usually caused by RNA viruses of the families Flaviviridae, Filoviridae, Arenaviridae and Bunyaviridae (4, 5). Almost similar severe outbreaks of hemorrhagic fever with high fatality rate occurred in the provinces of Zaire and Sudan. Although the causing agents were identical and similar in morphology to Marburg virus strain isolated from South Africa in 1975 and Germany in 1967 and was named Ebola virus, the electron microscopy and serological studies showed that the involved viruses were closely related (6). Zaire Ebola virus (ZEBOV) has the highest case fatality rates approximately 88% and death rate of 53% since its discovery (7). Ebola virus is mostly thrived in West and Black Africa where it has caused large outbreaks with high mortality rates. In spite of such severity of Ebola outbreaks, no affective vaccine or therapeutic drug is developed yet for clinical purposes.
2. Origin and Distribution
Five species of genus Ebola virus are identified, among which four species of Zaire Ebola virus (ZEBOV), Sudan Ebola virus (SEBOV), Bundibugyo Ebola virus (BEBOV) and Tai Forest virus (TAFV) are infectious for humans and are recognized from Africa; while one Reston Ebola virus (REBOV) characterized from Philippines is associated with nonhuman primates and pigs. This strain is considered as not being pathogenic for humans but causes hemorrhagic fever in experimentally infected animals (8, 9). It is said that the current outbreak of Ebola virus disease in West Africa found its roots from a single zoonotic transmission event by a two year old boy in Meliandou, Guinea (10).
3. Hosts
Natural reservoir for Ebola is not confirmed yet. However researchers suspect the fruit bats as the most probable candidate species. Three different bat types are found to carry this virus without being affected that suggested them as a primary natural reservoir for Ebola viruses (11). However infectious Ebola viruses have never been characterized from any fruit bat species except small amount of viral RNA fragments of Zaire Ebola virus around endemic areas (12-14). Birds, plants, and arthropods are also regarded as possible reservoirs of this virus. On the other hand several strains of filovirus are detected through enzyme-linked immunosorbent assay (ELISA) from the serum of migratory fruit bats (Eidolon helvum) in Zambia (14). EBOV-GP interactions with human and African fruit bats were compared, GP displayed similar biological properties in bats and human cells. Thus, results suggested that both have similar interactions with EBOV-GP (15). Insectivorous free-tailed bats (Mops condylurus) may be the potential source of African outbreak since it survived experimental infections, and index case may be infected by playing near the housing colony of these bats (10). Phylogenetic and sequencing evidence revealed that filovirus-like elements integrated in the genomes of bats, shrews, tenrecs, rodents and marsupials. This shows that the mammal-filovirus association is ancient which resulted in gene production (RNA or protein) (16).
4. Genome and Structure
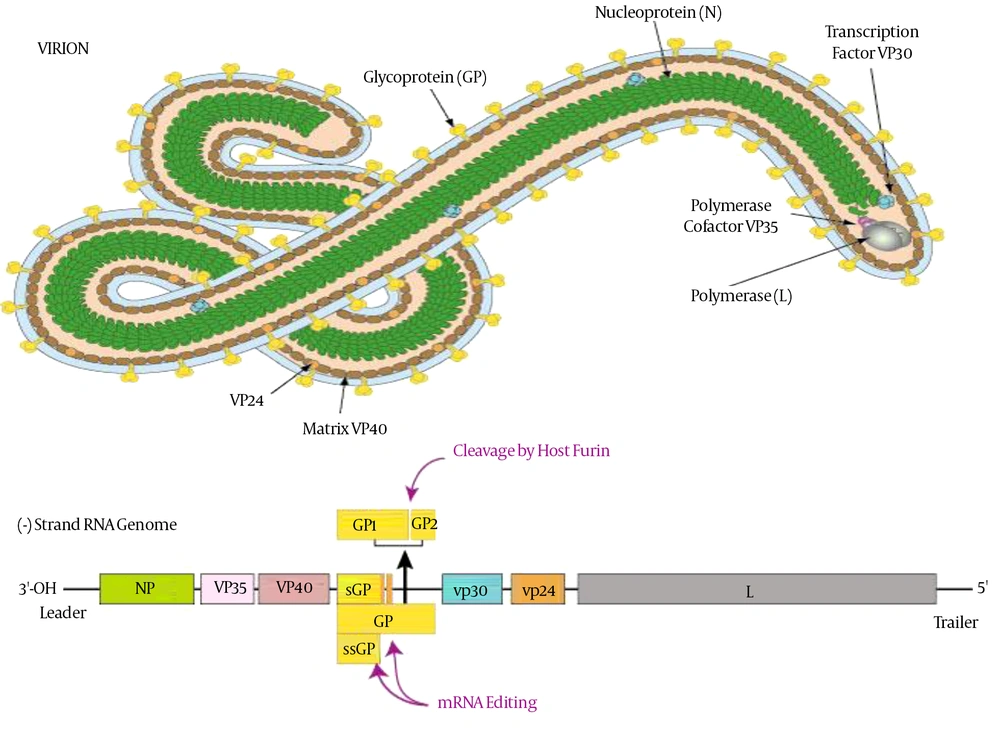
The Ebola virus (EBOV) particles contain a single negative-strand, ~19 kb, 18,959 to 18,961 nucleotides in length and helically wound RNA genome with seven linearly arranged genes encodes seven structural proteins. Four of these proteins nucleoprotein (NP), VP30, VP35 and L, are required for the transcription and replication of the viral genomic RNA by constituting a helical nucleocapsid (17, 18). Other three proteins are glycoprotein (GP), VP24 and VP40 are associated with viral membrane to form the filamentous virions (19, 20). Expression of the virion-associated proteins VP24 and VP35 led to assembly of nucleocapsids by transmission of electron microscopy and showed that it is involved in the assembly of EBOV nucleocapsids (21). The matrix protein VP24 is also involved in the regulatory process of viral genome replication and transcription (22). About 731 nucleotides from the 5' end and 472 nucleotides from the 3' end are enough for viral genome replication though not for infection. As Ebola virus is RNA coded, therefore it was found to mutate very rapidly within the host and reservoir population. The observed mutational rate of Ebola virus is 2.0 x 10-3 substitutions per site per year that is as fast as seasonal influenza. That is why it is very difficult to develop vaccine against Ebola virus. The structure of Ebola is cylindrical/tubular that contains viral envelope, nucleocapsid and matrix components. The diameter of cylinders is approximately 80 nm and the length may be 14000 nm having a spike like virally encoded glycoprotein (GP) of 7 - 10 nm long projects from its surface of lipid bilayer (23). The overall shape of virions varies considerably ranging from simple cylinders to branches, loops and reverse direction. However, the characteristic threadlike structure is a more general shape of filoviruses (Figure 1).
Structure and the Genome of Ebola Virus (24)
5. EBOV Receptors and Entry Mechanism
The EBOV entry process into the host cell consists of three sequential steps such as attachment, co-receptor binding and then fusion. For viral fusion, viruses mostly exploit the endocytic routes and through that, they access cytoplasm by macropinosomes, caveolae and clathrin-coated vesicles pathways. Different types of host cell-surface receptors are identified to play a role in EBOV entry into the host cell that are the β1 integrin receptors (25), galactose- and N-acetylgalactosamine-specific C-type lectin (hMGL) (26), dendritic-cell-specific intercellular adhesion molecules DC-SIGN-related (DC-SIGNR) and (ICAM)-3-grabbing nonintegrin (DC-SIGN) factors (27, 28). The EBOV also exploits several molecules such as a variety of different C-type lectins to entry the host cells (26).
6. Replication and Pathogenicity of Ebola Virus
Zaire Ebolavirus (ZEBOV) similar to Reston Ebolavirus (REBOV) can replicate and induce infection in pigs and can be transmitted to other animals. Experiments were conducted to evaluate the virus transmission from infected to naïve animals and results suggested that the virus replicated in the respiratory tract and transmission was confirmed in all naive pigs (29). EBOV proteins interact with various host proteins for virus replication. Several host proteins, such as Nieman-Pick C1, mucin domain 1, T-cell immunoglobulin, Tyro3 receptor tyrosine kinase family Axl, cathepsin L/B, Mer and Dtk were identified as important cellular factors involved in the EBOV entry step (30-34). The first cellular factor that contributes to the transcription and replication of the EBOV genome is DNA topoisomerase 1 (TOP1) but mostly the cellular host factors involved in EBOV transcription and replication are still unknown (35).
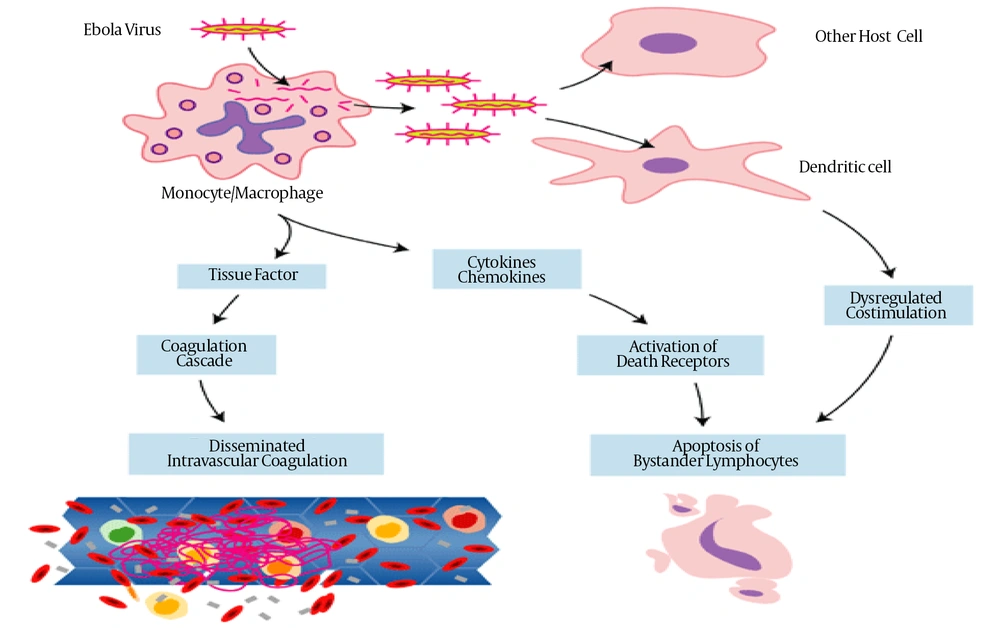
Paradigm of Key Events in Ebola Virus Pathogenesis in Primates (36)
7. Mode of Action
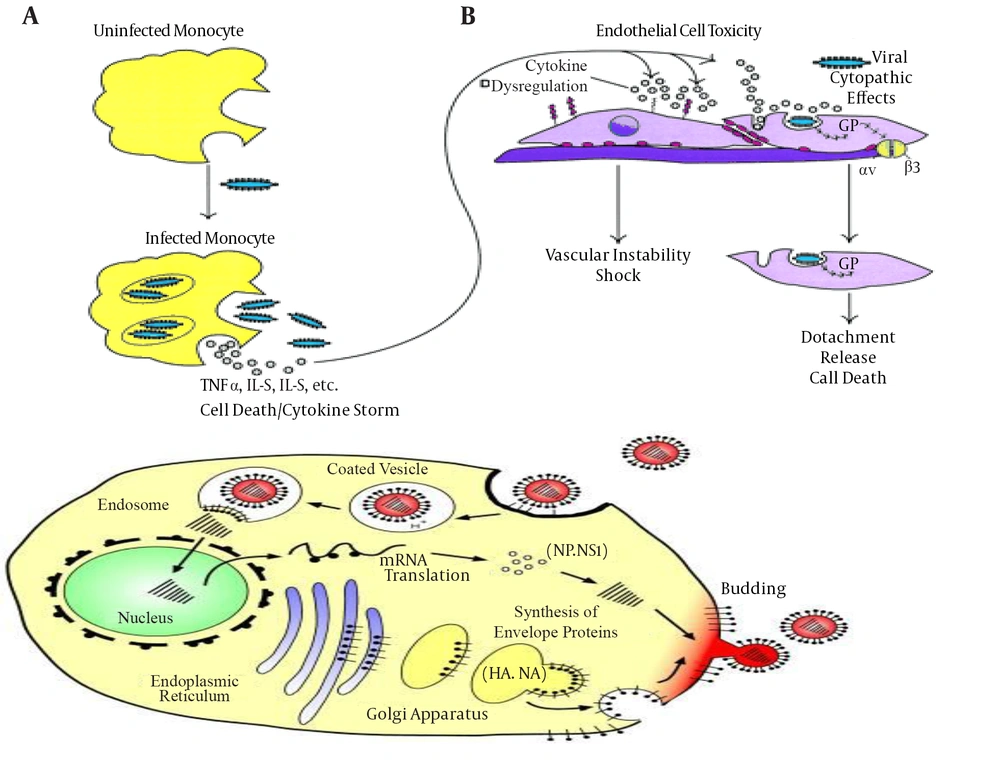
The EBOV-glycoprotein (EBOV-GP) has a molecular mass of approximately 140 kDa, mediates viral entry and promotes viral release from host cells (37). EBOV-GP is synthesized as single-chain precursors and further transferred co-translationally into the lumen of the endoplasmic reticulum to form trimmers (38). The receptor binding subunit of GP should be post-translationally cleaved into two chains that are GP1 and GP2, in order to enter and cause infection to proceed (39). Entry of the virus is pH dependent and also associated with the cleavage of GP by proteases (40). Cleavage produced three cleavage fragments, with masses of 23, 19, and 4 kDa (41). A 19-kDa core subunit triggers GP to bind endosomal receptors and potentiates GP to undergo subsequent fusion-relevant conformational changes (42, 43). The GP1 subunit is responsible for receptor binding while GP2 subunit anchors the glycoprotein into the viral membrane. The first 300 residues of GP1 subunit are conserved but the remaining residues are variable. The variable region of GP1 that is C-terminal, contains several O-linked glycosylation sites and is known as the mucin domain (44, 45).
The structure of GP2 by X-ray crystallography revealed that GP2 contains a central triple stranded coiled coil followed by a disulfide-bonded loop. GP2 could bridge two membranes to initiate membrane fusion (46). Thetrimeric crystal structure of surface glycoprotein (GP) is also required to initiate attachment and fusion of viral and host membranes that further bind to a neutralizing antibody, KZ52, in humans (42). Human lysosomal cholesterol transporter Niemann-Pick C1 (NPC1) fulfills a cardinal property of viral receptors and binds specifically to viral GP (30, 31). EBOV-GP is the main viral determinant of the Ebola virus pathogenicity that induces cytotoxic effects in human endothelial cells (47). GP contains a mucin-like domain involved in massive endothelial cell loss within 24 hours and resulted in increased vascular permeability into explanted human or porcine blood vessels. The mucin-like domain of GP was required for this effect and indicated that it was the viral determinant of Ebola pathogenicity (47). Another factor of AMPK in macropinocytic events was also needed for EBOV GP-dependent entry (48). These filoviruses maintain glycoprotein glycosylation to protect against antibody neutralization and proteases at the expense of efficient entry (3). Enzymes such ascathepsin B (CatB) and cathepsin L (CatL) play an essential role in diminishing the multiplication of infectious ZEBOV by carrying out proteolysis of the EBOV GP subunit GP1 (49).
Infection and Mode of Action of Ebola Virus (36)
8. Disease and Symptoms
Ebola virus has an incubation period of 2 to 21 days and during these days infected patients develop flu-like symptoms such as fever, headache, reduced appetite, vomiting, abdominal pain, vascular dysfunction, diarrhea and fatigue. Fever remains higher than 38.3°C during these conditions. These conditions are followed by vomiting, abdominal pain and sometimes chest pain and shortness of breath may occur. Severe and fatal stages are accompanied by bleeding from small infections, hemorrhagic diathesis, reduced adaptive immune responses, shock, coagulation disorders and eventual multiple-organ failure occur in this disease (50, 51). The fatal outcome is also associated with the aberrant innate immune responses due to massive intravascular B- and T-lymphocyte apoptosis that induced profound suppression of adaptive immunity in experimentally infected animals and humans (51, 52). Recovery may start 7 to 14 days after the first symptom (52). Death often occurs due to decreased blood pressure because of severe bleeding and loss of body fluid (53). Those who survive have many ongoing problems such as liver inflammation, muscular and joint pain, decreased hearing, weight loss and weakness. Recovered patient can no longer transfer the virus (54).
9. Epidemiology and Transmission of Ebola
High numbers of Ebola virus particles can be found in sweat glands of the human skin indicating that transmission may occur through direct contact but viral entry in the body is still unclear. Human-to-human transmission mainly occurs via contact with the body fluids of the infected person during the treatment reusing unsterilized medical devices and traditional funeral practicing, kissing, touching and washing the body (55). Airborne mode of transmission between primates including humans is not observed yet either in natural conditions or in laboratories. The transmissibility of Ebolavirus is generally highest in the clinical course of infection unlike other viruses (56). This virus may reach any place in the world with no time through modern transportation. Investigation suggested that environmental factors are also associated with transmission of Ebola virus disease (EVD) particularly to the drier conditions at the end of the rainy season and may enhance transmission of Ebola virus from its cryptic reservoir to humans (56).
10. Historical Outbreaks and the Studied Cases
Ebola virus disseminates in Black Africa, where it often drives large outbreaks of acute hemorrhagic fever with high fatality rate (55). The first case of filovirus hemorrhagic fever was reported in 1967 in Germany and the former Yugoslavia, and the causative agent was identified as Marburg virus. After that, 12 similar cases of hemorrhagic fever were described in 1976 from outbreaks in two neighboring locations: first in southern Sudan and subsequently in northern Zaire, now Democratic Republic of the Congo (DRC). This strain caused three more epidemics in the areas of Nezara, Sudan in 1979 (57). An unknown causative agent was isolated from patients in both outbreaks and named Ebola virus after a small river in northwestern DRC. These two epidemics were caused by two distinct species of Ebola virus, Sudan Ebola virus (SEBOV) and Zaire Ebola virus (ZEBOV) (58). Outbreaks of ZEBOV, SEBOV and BEBOV hemorrhagic fever occur intermittently in Africa and are associated with more than 90% of case-fatality rates (15). Tai Forest virus (TAFV) was observed in an infected ethnologist who used to work in the Tai Forest in Côte d’Ivoire in 1994 in Africa. Later it was identified that this newly discovered virus had the same aspect of Ebola hemorrhagic fever as the other two Ebola strains (59). In 1994 an epidemic was primarily reported as a yellow fever outbreak in 44 cases with 28 deaths in Gabon. Later studies suggested that it was due to Ebola virus attack.
Gabon was attacked twice by Ebola virus in 1996. The first outbreak began in February and out of the 37 cases 21 died. The second epidemic was from July to December and 40 deaths were reported among the 52 identified cases (60). Ebola hemorrhagic fever was re-emerged in 1995 in Kikwit, Democratic Republic of the Congo. Isolates obtained from the samplings confirmed the presence of Zaire Ebola Virus (ZEBOV); viruses that caused severe hemorrhagic fever outbreak among the people of Kikwit, Democratic Republic of the Congo in 1995 and 231 people died out of 300 affected people. Similar isolates were also observed during an outbreak of this fever in cynomolgus macaques in Texas, Alice and the Philippines in 1996 (46, 61). The 2nd outbreak of Sudan Ebola virus was observed in areas of Gulu, Mbarara, Masindi and Uganda in 2000. With a total number of about 430 cases reported in Uganda in 2000-2001, this Ebola virus outbreak is considered as the largest epidemic described to date. The 3rd Sudan Ebola virus (SEBOV) epidemic was observed in 2004 in Yambio and again in Sudan (57).
A novel strain of Ebola virus named Bundibugyo ebolavirus was identified during an outbreak of viral fever in Bundibugyo district of western Uganda and later on in equatorial regions of Africa in 2007-2008. The Bundibugyo Ebola virus outbreak caused a lower proportion of deaths than did the Sudan Ebola virus outbreak in Gulu and Sudan. Uganda ministry of health (UMOH) declared deaths of 39 persons in this outbreak in Bundibugyo district (62). In the end of August 2014, World Health Organization (WHO) was notified about another severe outbreak with similar observations of Ebola viral disease with 74% death rate in the locality of Boende town, Equator province of western democratic republic of Congo (DRC). Fatality rate was 74%. It was the seventh Ebola outbreak which occurred in the DRC with the first in 1976 in Sudan and Zaire (63). Before 2014, approximately more than 2400 people were infected, along with more than 1500 recorded deaths in the past four decades since its first discovery in 1976 in the areas of Zaire and Sudan (64). From December 2013 to September 2014, a total of 2296 deaths were reported out of 4507 confirmed and probable cases of Ebola virus from five countries of West Africa i e, Guinea, Nigeria, Senegal, Liberia, and Sierra Leone (65). The largest outbreak is the ongoing epidemic in some specific areas of West Africa including main targets of Guinea and Sierra Leone. WHO reported a total of 28,599 cases observed and 11,289 deaths from this outbreak by 10 November 2015 (66). There were about 24 outbreaks of EVD, but only seven involved more than 100 cases. There were 15 generations of viral transmission in 1976 SUDV outbreak in Southern Sudan with 284 cases and 53% mortality while the outbreak of four generations of EBOV in northern Zaire, and DRC had 315 cases and 88% mortality rate (67).
11. Countries with Widespread Transmission and Current Scenario
According to the world health organization (WHO), about 24 outbreaks of Ebola virus disease (EVD) were recognized. Between the first recognized outbreak in 1976 by SUDV and onset of 2013 - 15 in West Africa by EBOV, about 3,400 cases were reported.
| Year | Species | Country | Cases | Deaths | Case Fatality, % | References |
|---|---|---|---|---|---|---|
| 2014 - 15 (Sep - Sep) | ZEBOV | Guinea | 3805 | 2536 | 67 | (66) |
| 2014 - 15 (Sep - Aug) | ZEBOV | Liberia | 10672 | 4808 | 45 | (66) |
| 2014 - 15 (Sep - Sep) | ZEBOV | Sierra Leone | 14122 | 3955 | 28 | (66) |
| 2014 - 15 (Sep - Nov) | ZEBOV | All countries | 28599 | 11289 | 39 | (66) |
| 2014 (July - Oct) | ZEBOV | Africa (DRC) | 69 | 49 | 71 | (63) |
| 2014 (Jan - Sep) | ZEBOV | Guinea,West Africa | 4507 | 2296 | 70.8 | (65) |
| 2012 | BEBOV | Africa (DRC) | 57 | 29 | 51 | (68) |
| 2012 (July - Aug) | SEBOV | Kibaale, Uganda | 11 | 4 | 36 | (68) |
| 2012 Nov | SEBOV | Luwero, Uganda | 6 | 3 | 50 | (68) |
| 2011 | SEBOV | Uganda | 1 | 1 | 100 | (69) |
| 2008 | ZEBOV | Africa (DRC) | 32 | 14 | 44 | (70) |
| 2007 | BEBOV | Uganda | 149 | 37 | 25 | (71, 72) |
| 2007 | ZEBOV | Africa (DRC) | 264 | 187 | 71 | (73) |
| 2005 | ZEBOV | Africa (DRC) | 12 | 10 | 83 | (74) |
| 2004 | ZEBOV | Africa (DRC) | 35 | 29 | 83 | (13) |
| 2004 | SEBOV | Sudan | 17 | 7 | 41 | (75) |
| 2003 (Nov - Dec) | ZEBOV | Africa (DRC) | 35 | 29 | 83 | (76) |
| 2003 (Jan - Apr) | ZEBOV | Africa (DRC) | 143 | 128 | 90 | (76) |
| 2001 - 02 | ZEBOV | Africa (DRC) | 59 | 44 | 75 | (76) |
| 2001 - 02 | ZEBOV | Gabon | 65 | 53 | 82 | (76, 77) |
| 2000 | SEBOV | Uganda | 425 | 224 | 53 | (78) |
| 1996 (Jul - Dec) | ZEBOV | Gabon | 60 | 45 | 75 | (79) |
| 1996 (Jan - Apr) | ZEBOV | Gabon | 31 | 21 | 68 | (79) |
| 1995 | ZEBOV | Africa (DRC) | 315 | 254 | 81 | (67, 80) |
| 1994 | TAFV | Cote d'Ivoire | 1 | 0 | 0 | (59) |
| 1994 | ZEBOV | Gabon | 52 | 31 | 60 | (79) |
| 1979 | SEBOV | Sudan | 34 | 22 | 65 | (81) |
| 1977 | ZEBOV | Africa (DRC) | 1 | 1 | 100 | (82) |
| 1976 | SEBOV | Sudan | 284 | 151 | 53 | (83) |
| 1976 | ZEBOV | Africa (DRC) | 318 | 280 | 88 | (83) |
12. Diagnostic Strategies
It is very important to recognize Ebola virus infections quickly to limit the spread of the disease further. For this purpose, various diagnostic techniques and assays are employed to detect the virus type in the body including transmission electron microscopy (TEM), virus culture, immunohistochemistry, conventional reverse transcription-PCR (RT-PCR), antigen and antibody detection ELISAs etc. (61, 84-86). A Fluorogenic 5’ nuclease assay having one-tube reverse transcription-PCR was made and tested by the ABI PRISM 7700 sequence detection system that consisted of one common primer set and two differentially labeled fluorescent probes. This assay can detect and differentiate two subtypes of Ebola virus such as ZEBOV and SEBOV simultaneously (87).
13. Clinical Implications or Applications
There were a number of efforts in clinical settings to recover the status of EBVD infected patients, and several clinical diagnostic strategies and therapies were developed in animal models. Research work on filoviruses is difficult because these viruses require special containment for safe research due to Biosafety Level 4 (88). Various antiviral and modulated host immune response strategies can be deployed to treat patients.
14. Disinfection
Ebola virus is susceptible to glutaraldehyde, sodium hypochlorite, 3% acetic acid (pH 2.5), β- propiolactone, paraformaldehyde and formaldehyde. Various dilutions of sodium hypochlorite may be used to disinfect Ebola virus. Various other chemicals such as Calcium hypochlorite, ether, methyl alcohol, peracetic acid and sodium deoxycholate are also effective against filoviruses. These viruses can be also inactivated by gamma irradiation, ultraviolet light, boiling for 5 minutes or heating to 60°C for 30 - 60 minutes (89).
15. Vaccination and Medication
Marburg and Ebola filoviruses caused infectious hemorrhagic fevers and resulted in up to 90% human mortality rates while there is no useful vaccine or therapeutics available for clinical purposes. The highly lethal and infectious nature of these viruses stresses the need for reliable diagnostic methods. The repeated and continued outbreaks of filoviruses are major concerns from the public health and biodefense perspectives since no effective treatment is available (90). The complete eradication of smallpox from the globe was a fact that triggered optimism about the feasibility to eradicate all major infectious diseases of the mankind. Due to lack of licensed therapeutic treatment, non-pharmaceutical interventions such as case isolation, sanitary burial, contact precautions and other quarantine practices should be relied upon to curb transmission. Scientists are trying to hail vaccines against killer Ebola virus and most of them are on the stage of clinical trials. Several candidate vaccines such as bivalent RABV/ZEBOV vaccine expressing EBOV-GP by a reverse genetics system (91),Virus-like particle (VLP)-based vaccines against EBOV in nonhuman primates known as Ebola VLPs (eVLPs) (92), panfilovirus vaccine based on a complex adenovirus (CAdVax) technology (93) etc. are tested. Replication-competent vaccines based on attenuated recombinant vesicular stomatitis virus vectors against EBOV and MARV elicited completely protective immune responses in nonhuman primates (66, 94).
16. Prevention
In the present circumstances, the only thing that can save us from this virus is prevention. People who are associated with and dealing with Ebola hemorrhagic disease should wear protective clothing with no skin exposure, recommended by US centers for disease control. These measures should be adopted when handling the contaminated objects and instruments. Educating the general public about the Ebola infectious risks and protective measures to prevent infection is recommended by the WHO. Direct contact with the dead body should be avoided. Social anthropologists can help with finding alternatives to traditional rules for burials. Bio-safety level 4 is required in the laboratories where diagnostic testing is carried out and lab attendants must be also trained in BSL-4 practice (95).
17. Future Thrusts and Strategies
Ebola virus can be potentially used as a biological weapon against enemies but it might be difficult to prepare it for mass destruction because of its in-affectivity in open air (24, 96). One example of its use as bio-weapon is that the British broadcast channel (BBC) reported in 2015 that North Korean state media suggested the disease was created by the US military as a biological weapon (95). It should and must startle the will of international society to cooperate and act upon. Projections as of all previous outbreaks showed that, with this exponential growth rate and no immediate care, accumulative infections of Ebola could rise to a record break number in upcoming years. Currently, patients are sent home with no cure, which shows no chance for control efforts. Highly lethal and infectious nature of this virus stresses the need for reliable diagnostic methods (96). Now it is time to show the utmost commitment to respond to this natural disaster, its present and future impact.