1. Context
According to the 3rd edition of the International Classification of Diseases for Oncology, tumors of the central nervous system (CNS) are those affecting the spinal cord and brain, including the pituitary gland, meninges, pineal gland, and nerves (1). Brain tumors have been traditionally classified based on the microscopic investigation of hematoxylin and eosin- (H & E) stained tissue sections. The increased comprehensive knowledge in relevant genetic alterations and mutations with clinical outcomes resulted in the incorporation of molecular signatures as a part of the diagnosis, management, and treatment of CNS malignancies (2). Various brain cancer susceptibility genes are involved in DNA damage response, which strongly indicates that critical DNA repair pathways and checkpoint controls are necessary for preventing tumor malignancy (3). Since the number of prognostic and predictive neuro-oncologic genetic markers is steadily increasing, comprehensive analyses of the molecular techniques used to examine neuro-oncology samples are vastly required for the evaluation of brain tumor specimens in a modern pathology setting. Molecular analysis and profiling of brain cancers lead to improved diagnostic accuracy, target identification and predictive prognosis (4). The recent development of NGS technology and other complementary genomic platforms have transformed our capacity to investigate the molecular landscapes of human cancers, including brain tumors (5). In this review, we summarized and analyzed the results of high-throughput genomic studies, such as NGS, in order to diagnose various types of brain tumors.
2. Gliomas
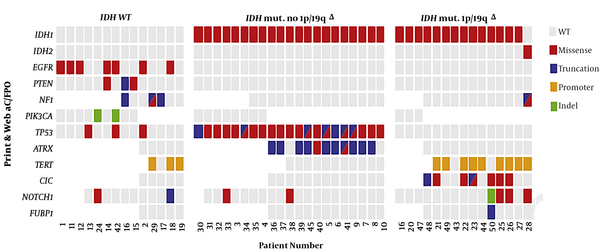
Diffuse gliomas (DGs), the most common primary malignancy of the CNS, generally affect the cerebral hemispheres of adults. According to the World Health Organization (WHO) classification of the CNS tumors, lower-grade gliomas or grades I and II DGs are oligodendrogliomas, astrocytomas, and oligoastrocytomathat the latest is a less well-defined type of oligodendroglial (OA) (6). It has been reported that molecular landscape, such as 1p/19q co-deletion status, is a better predictor of clinical outcome than that of the histologic distinction between oligodendroglioma and OA (7). Therefore, the WHO has recently updated the CNS tumor classification (8). In fact, NGS is an effective technique for the detection of a wide variety of molecular alterations, which provides an efficient, attractive, as well as a cost-effective unimodal molecular platform for the classification of lower-grade gliomas based on the 2016 WHO CNS criteria (9, 10). The results of the sequencing of the samples obtained from patients with lower-grade gliomas, which were organized by IDH1, IDH2, and NGS-based 1p/19q co-deletion status, are shown in Figure 1 (11). Carter et al. have used a single, targeted NGS panel to detect single-nucleotide variation and copy number variation in 50 patients with lower-grade gliomas, including 36 oligoastrocytomas, 11 oligodendrogliomas, 2 astrocytomas, and 1 lower-grade gliomas. They have found that NGS is superior to fluorescence in situ hybridization (FISH) and immunohistochemistry in the detection of segmental chromosomal mutation from whole-arm deletions in glioma tumors (11). Targeted NGS for mutations in TP53, ATRX, CIC, IDH1, IDH2, FUBP1, PI3KC, EGFR, H3F3A, TERT, BRAF, PTEN, and NOTCH genes and for copy number alterations of chromosomes 1p, 19q, 10q, and 7 has been also used to classify DG (12) . Indeed, DGs could be classified as type I (1p/19q codeletion, “oligodendroglioma”), type II (IDH mutation, “astrocytoma”), type III (TERT mutation or 7+/10q and 1p/19q intact, “glioblastoma”), and childhood glioblastoma (H3F3A mutation) (12).
The results of the sequencing of the samples obtained from patients with lower-grade gliomas were organized by IDH1, IDH2, and NGS-based 1p/19q co-deletion status. The presence of more than one variant is shown in multiple color boxes. WT, wild type; mut, mutant (Published by Carter et al. (11)).
3. Pilocytic Astrocytoma
Pilocytic astrocytomas (PAs) are WHO grade I CNS tumors that are most common in children with the age of less than 19 years old (0.84 per 100,000) with approximately 700 new cases being annually diagnosed in the United States (13). Despite the WHO classification, these tumors are still clinically referred to by other terms, including optic glioma, cerebellar astrocytoma, and infundibuloma due to their certain anatomic sites and the distinct predilection in young patients (14). Considering the difficulty in diagnosing PA using anaplastic features that are exclusively based on morphology, applying the relevant molecular biomarkers to diagnose the presence of mitogen-activating protein kinase (MAPK) pathway alterations are necessary (15). The results of whole genome sequencing have shown that single alterations of the MAPK pathway are solely detected in almost all PA cases, indicating that PA represents a one-pathway disease (14). Activation of MAPK signaling through various described mutations or alterations, the commonest tandem duplication is located at 7q34 that leads to KIAA1459-BRAF fusion, is present in most of the PA cases. Therefore, the identification of a known mutation/alteration is useful to support the diagnosis of PA (16-18). To investigate the full range of genetic alterations present in PA, the whole genome sequencing of DNA obtained from blood has been performed. Jones et al. have identified KIAA1549:BRAF fusion variants, FAM131B:BRAF fusion, four BRAFV600E mutations, and one BRAFins599T in pediatric brain tumors by NGS analysis (16). It has been also detected a new recurrent alteration, namely mutation of two hotspot residues (N546 and K656) in the kinase domain of FGFR1 that are occasionally observed in adult glioblastoma (GBM) (19). The NGS analysis has also demonstrated that GIT2-BRAF fusion is identical to that detected in other fusion alterations in PA. This fusion most likely acts as a tumor driver that activates the MAPK signaling pathway (20).
4. Glioblastoma (GB)
Glioblastoma multiforme (GBM) is an extremely malignant CNS tumor. Mutation and alteration of the epidermal growth factor receptor (EGFR) have been frequently reported in various human malignancies, such as GBM (21). However, the presence of multiple types or heterogeneous mutations is one of the GBM hallmarks that leads to insufficient single-agent activity over standard treatments in patients with GBM (22). Therefore, the determination of predictive biomarkers can predict the fate of EGFR-targeted therapy that resulted in help to the patients with GBM and improved the treatment sensitivity. In addition, detailed mutational analyses of brain tumors may reveal the mechanisms underlying drug resistance that may finally enhance the effectiveness of personalized cancer therapy (23). Spatial heterogeneity in PDGFRA, TP53, and EGFR genes in glial tumors has been previously identified (22). Furthermore, the results of NGS analyses have demonstrated the presence of FLNA point mutation, p.V120M and EGFR mutations, p.A289V pP772 delinsPPl, in the GBM patients (24). Both MutSig and InVEx algorithms have been previously identified as mutant genes in GBM patients, namely TP53, EGFR, PTEN, PIK3CA, NF1, RB1, PIK3R1, IDH1, leucine-zipper-like transcriptional regulator 1 (LZTR1) and PDGFRA (25). In addition to the alterations found in the signature oncogenes of GBM, NGS analysis has revealed that more than 40% of GBM neoplasms harbor at least one non-synonymous mutation among the chromatin-modifier genes. Somatic alterations analysis by NGS confirmed mutual exclusivity among alterations affecting Rb pathway (CDK4, CDK6, CCND2, CDKN2A/B and RB1), p53 pathway (MDM2, MDM4 and TP53), and different components influencing PI3K pathway (NF1, PIK3R1, PIK3CA, EGFR, PTEN, and PDGFRA) (25). Ballester et al. (2) have studied mutations of cancer-associated genes in 381 patients with brain tumor using commercially available NGS panels. They have found that 32% of cases with anaplastic ependymoma, atypical choroid plexus papilloma, and anaplastic meningioma showed no mutations in any of the regions of PNET, and DNET examined by NGS assay; similar to those detected in 37% of glioblastomas, 85.7% of ependymomas, and 100% of medulloblastomas. While, the most commonly mutated genes in brain tumors were TP53, IDH1, PIK3CA, PTEN, and EGFR with a frequency of 37.2%, 29.4%, 8%, 8%, and 7.5%, respectively (2). The most commonly mutated genes detected in GBM patients are TP53, IDH1, PTEN, EGFR, and PIK3CA with a frequency of 35.9%, 14.6%, 10.8%, 9.2%, and 8.4%, respectively. Moreover, the majority of GB and astrocytomas cases with one IDH1 alteration or mutation also have a TP53 mutation (87.5% and 84.9%, respectively). In grade II/III or astrocytomas, the frequency of TP53 and IDH1 mutations was significantly higher than other gene mutations, 59.0% and 63.8%, respectively. Moreover, PTEN mutations have been reported to be more common in GB than in lower-grade astrocytomas II/III. The most common mutations detected in anaplastic oligodendrogliomas are IDH1 (76.19%), followed by TP53 (23.8%), and PIK3CA (19.0%). In oligodendrogliomas (WHO grade II) the most common mutation are IDH1 (81.8%), PIK3CA (9%), and NRAS (9%) (2). Genetic analysis has been shown that recurrent mutation in IDH1 gene in most GBM cells. Deng et al. (26) have reported that the patients with grade IV primary GBM exhibited mutations in IDH1 and IDH2 with a frequency of 6.98% and 4.65%, respectively. They have reported that the frequencies of IDH1 and IDH2 mutation in grades II, II-III and III of brain glioma tumors are higher than those of grades I and IV (26).
However, several retrospective clinical trial studies have revealed the presence of wild-type IDH1 in GBM with bad prognostic outcomes compared to IDH1 mutation. The prognostic importance of IDH1 gene mutation has been recently identified in patients with GBM. Multiple studies have reported that the mutation of IDH1 acts as an important and independent factor for the progression-free survival as well as predicting longer overall survival in patients with GBM (27). In addition, the recent meta-analysis study has shown that GBM patients with epigenetic mutations such as MGMT methylation are also associated with longer overall survival not in progression-free survival (28).
5. Ependymoma
Ependymomas originate from different regions of the CNS with histological similarities. However, these tumors possess site-specific prognoses as well as genetic and transcriptional profiles alterations (29, 30) Nine supratentorial ependymomas (SE) have been recently analyzed by whole genome sequencing and it has been revealed that more than two-thirds of the SE tumors contain oncogenic fusions between RelA, the canonical NF-kB signaling member, and an uncharacterized gene, C11orf95 (31). These data have demonstrated that genetic alterations of RelA in human cancer are highly recurrent. In addition, the RelA-C11orf95 fusion protein can be a potential therapeutic target in the SE (31). Structural variations within chromosome 11 (11q12.1 and 11q13.3), producing catastrophic disruption of the locus, have been also detected by NGS in patients with SE (31).
6. Medulloblastoma (MB)
Medulloblastoma (MB) is the commonest primary malignant brain tumor in pediatric patients. Overall survival rates for patients with MB have reached 70% - 80% using the combination of surgery, craniospinal radiotherapy, and chemotherapy (32). The current consensus is that MB consists of four subgroups, including WNT, SHH, groups 1 to 4 that exhibit highly disparate mutational spectra, cytogenetics, and gene expression signatures (33). Medulloblastoma has been now considered four distinct diseases, each of which requires distinct therapeutic approaches (33).
The WNT subgroup of MB often carries heterozygous TP53 mutations and harbor nearly ubiquitous CTNNB1 mutations (34). The WNT subgroup of MB exhibits largely balanced genomes, with the exception of monosomy 6 that is a hallmark chromosomal aberration detected in almost all cases with WNT subgroup of MB; however; it is very rarely reported in other MB subgroups (35). Germline mutations that affect negative regulators of the SHH signaling pathway, either PTCH1 or suppressor of fused homologue (SUFU), induce the development of SHH-driven MB (36). The genome of the SHH subgroup of MB contains more remarkable chromosomal loss and gain regions such as partial deletions of chromosomes 9q and 10q as well as other less common aberrations than that of WNT subgroup of MB (37). In addition, MB tumors harboring mutations in the key SHH pathway target genes (i.e. GLI1 and 2, or MYCN) may have an inherent primary resistance against antagonists of SHH pathway signaling (37). Amongst four subgroups of MBs, the worst therapeutic outcome belongs to patients with group 3 MB (37). The high-level of mutation/amplification of the MYC proto-oncogene is highly enriched in this subgroup so that all cases mostly exhibit aberrant MYC expression (38). This subtype also exhibits high levels of genomic instability and often harbors deletions of 17p and 10q, 11q, and 16q as well as gains of chromosomes 1q, 7q, and 17q (37).
The most common subgroup of MB is the group 4. Cyclin-dependent kinase 6 (CDK6) and MYCN proto-oncogenes are recurrently amplified in the group 4 medulloblastomas (38). Also, i(17q) has been found in the majority of cases with this subgroup and most female patients lose one copy of the X chromosome (35).
A significant discovery in the genetic landscape of MB was made in 2011, when the first unbiased whole exome sequencing study of medulloblastoma was published. Somatic mutations in mixed-lineage leukemia (MLL) 2, MLL3, and histone methyltransferases (HMTs), were identified in some of MB cases based on Sanger sequencing (39).
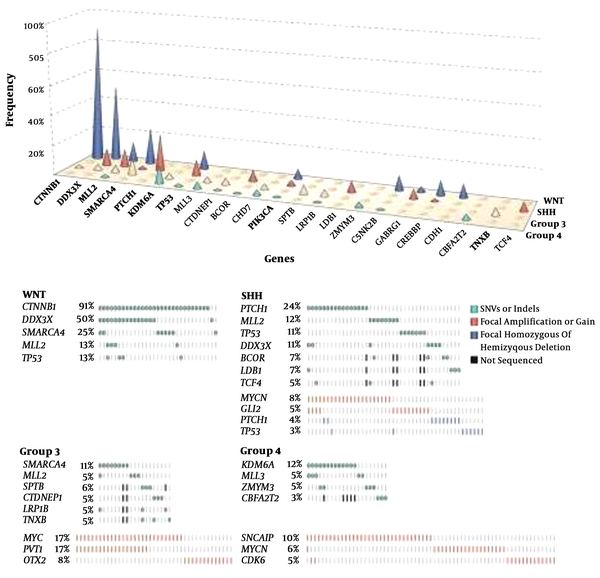
Next generation sequencing of all four subgroups of MB was published in a single issue of Nature (Figure 2).
Meta-analysis of MBNGS studies (published by Northcott et al (35)) is represented.
7. Neuroblastoma
Neuroblastoma (NB) is the most common solid tumor in children under the age of 10 years, contributing to approximately 15% of childhood cancer-related deaths (40). They are highly heterogeneous and represent a wide spectrum of clinical manifestations, as well as the likelihood of being cured, ranging from spontaneous regression to relentless progression despite multimodal treatments. These diversities in tumor behavior can be attributed to differences in the extensive underlying molecular mechanisms. Numerous genetic alterations are suggested (41). Germline mutations, namely anaplastic lymphoma kinase (ALK) and paired-like homeobox 2B (PHOX2B), mainly occur in familial NB (FNB). However, mutations in ALK gene, which lie in human chromosome 2p23, are more common than those in PHOX2B gene. In FNB, ALK mutations dominantly occur in coding areas, including hotspots R1275, F1245, and F1174 (42). Among these hotspot loci, not only the R1275Q mutation is the commonest germline ALK mutation, which occurs in hereditary NB, but also is the most common somatic ALK alteration (43).
Several common copy number variations [CNVs]) and genomic variables (single nucleotide polymorphisms [SNPs]) associated with sporadic NB have been identified using NGS. The first category of genomic variables was detected in high-risk patients with NB, including CNVs/14, BARD1, LMO1, LIN28B, and HACE1. While, low-risk gene mutations, such as DUSP12, DDX, IL31RA and HSD17B12 have been found in the second category. The variation of germline copy numbers, for instance, NBPF23 gene, has been detected in the third category (44-46). Common SNPs on chromosome 6 (6p22) within CASC14 and 15 genes, which are associated with NB risk, have been reported by Maris et al. (47). The relationship between invasive NB and several SNPs in BRCA1 associated with RING domain 1, which is located in the area 2q35 on human chromosome, has been also reported (48).
8. Meningiomas
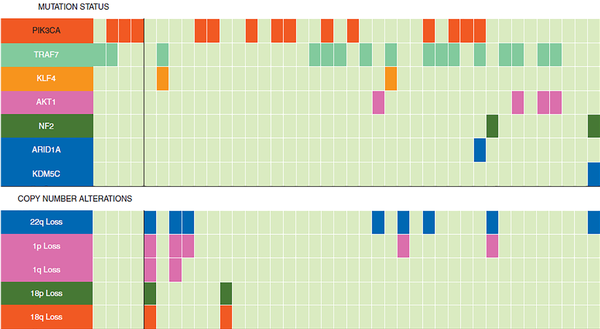
Meningiomas are the most reported primary tumors of the CNS in adults. All exons of known frequently mutated genes in these neoplasms, including ARID1A, AKT1, AKT3, ATRX, CHEK2, CREBBPKDM6A, KDM5C, NF2, PRKAR1A, KLF4, PIK3R1, PIK3CA, POLR2A, PDCD10, SMARCB1, PTEN, SMARCE1, SMO, SUFU, TERT, and TRAF7, have been identified using NGS assay (49). A genetic variation between progestin-associated meningiomas and non-hormonal treatment meningioma has been also reported. The two most frequent mutated genes in meninigioma are TRAF7 and NF2 (50). However, the higher frequency of PIK3CA and TRAF7 mutations and a lower frequency of NF2-related meningioma tumors have been reported post-long-term progestin therapy and targeted NGS (51). The mutational profile of progestin-associated meningiomas is illustrated in Figure 3 (51).
Mutation status and copy number alterations used for profiling progestin-associated meningiomas. The top and bottom panels display mutation status and copy number alterations, respectively (published by Peyre et al. (51)).
9. Astroblastomas
Astroblastoma is a rare and controversial primary brain tumor that is mostly diagnosed using histological approaches. This tumor typically displays indolent behavior with good prognosis; however, some of its variants are occasionally aggressive and infiltrative (52). The analysis of genomic alterations of astroblastoma has demonstrated that chromosomal gain is evident in arms of 20q, 9q, and gain of chromosome 19 as well as rare losses of chromosome 10 (52). Next-generation targeted exome sequencing of 300 cancer-related genes for astroblastoma was performed by Bale et al. (52). The NGS analyses of these genes in patients with astroblastoma have revealed multiple monosomies involving chromosome 10 (leading to single-copy loss of PTEN), polysomy of chromosomes 5, 9, 13 (resulting in single-copy loss of RB1), and 18, single-copy loss of 6q (including MYB and PARK2), and the loss of a region of one to two copies at 9p23-9p11.2, possibly leading to homozygous deletion of the tumor suppressor gene, CDKN2A. In addition, out of four cases with astroblastoma, two patients showed SETD2 variations (52).
10. Discussion
In this review study, we summarized and analyzed the results of high-throughput genomic studies, such as NGS, that can independently identify the integrated diagnosis of brain tumors, including glioblastoma, pilocytic astrocytomas, medulloblastomas, ependymomas, neuroblastoma, meningiomas, and astroblastoma across a variety of entities with pathognomonic genetic alterations. In adults, the most common primary brain tumors are gliomas, such as glioblastomas, astrocytomas, oligodendrogliomas and ependymomas, displaying variable biological behaviors. Approximately, 250,000 people worldwide are annually diagnosed with brain cancers (53). Human primary or intrinsic brain and CNS neoplasms indicate characteristic molecular signatures consistent with tumor type. Numerous studies have recently focused on analyzing genomic alterations in brain tumors. Detection of alterations or mutations in specific genes of some brain tumors, such as glioma, has revolutionized our understanding of the pathogenesis of many types of glioma (54).
The diagnosis, management, and treatment of patients with intrinsic or primary brain tumors have been previously dependent on a classification system using protein expression levels, microscopic and immunohistochemical examinations. The increased knowledge of relevant genetic alterations or genomic landscape of primary human brain tumors and clinical outcomes led to the incorporation of molecular signatures in the diagnosis, management, and treatment of brain malignancies (55). Based on the latest WHO classification of the CNS tumors (2016), molecular investigations of primary brain tumors have become an important part of the diagnostic workup of human CNS tumors (8). The advances in sequencing technologies have recently resulted in the incorporation of NGS assays in many clinical diagnostic laboratories and have increased the demand for identifying molecular profiles of human brain tumors (4). It has been demonstrated that there is a significant histological overlap between brain tumors, such as astroblastoma with GB, particularly in the absence of characteristic molecular signatures of the tumor (56).
In total, NGS is an attractive, efficient, and cost-effective technique in detecting a wide variety of molecular alterations, including genomic mutations such as insertions and deletions (indels), CNVs, single-nucleotide variations, and SNPs, which make it as a supplier unimodal molecular platform for the classification of human brain tumors.
Genomic characterization of brain neoplasms has been recently performed using NGS and has resulted in the generation of a large amount of information that can be very usual in the practice of neuropathology (4). The NGS analysis has shown that the most clinically relevant genes for brain neoplasms are TP53, IDH1, IDH2, PIK3CA, EGFR, BRAF, PDGFRA, and FGFR1, 2 and, 3. According to the 2016 CNS WHO guidelines, molecular testing of IDH1 and 2 genes are critical for the diagnosis and management of diffuse gliomas. As TP53 mutations are rare in neoplasms with 1p/19q co-deletion, TP53 can be helpful to identify DGs that are 1p/19q-intact (2). The commercial NGS assay serves to detect IDH-wild-type, IDH, and TP53-mutant status in diffuse glioma. The combination of a separate assay to identify 1p/19q status using NGS is also helpful for the molecular classification of most of gliomas such as GBM based on the latest WHO CNS tumor classification. The expression or genomic profiles of BRAF, PIK3CA, PDGFRA, EGFR, and FGFR1, 2 and 3 can be helpful to choose the most appropriate therapeutic approach (2).
11. Conclusions
This review study indicates that the NGS technique can refine or independently help in the integrated diagnosis of intrinsic primary human brain tumors across a variety of entities with pathognomonic genetic mutations. Furthermore, the utility of a limited, targeted, hybridization-capture-based NGS panel in identifying the relevant alterations is necessary for subtyping of gliomas using the recent WHO CNS tumor classification (updated 2016). It has been emphasized that relevant histopathological and molecular information is required to improve the accuracy of management and diagnosis of primary human brain tumors. The recent breakthroughs made in the field of brain tumor genomics and NGS studies may influence ongoing basic research and help to choose the most appropriate therapeutic approach.