1. Context
Several coronaviruses can infect humans to cause mild to fatal respiratory disease. Coronaviruses with the highest fatality risk include the Middle East respiratory syndrome coronavirus (MERS-CoV), severe acute respiratory syndrome coronavirus (SARS-CoV), and the novel coronavirus (2019 novel coronavirus, 2019-nCoV) that was initially characterized in Wuhan, China (1). The latter virus, which is currently named as SARS-CoV-2 by the International Committee of Taxonomy of Viruses (ICTV) (2), can cause coronavirus-associated acute respiratory disease, coronavirus disease 19 (COVID-19). Globally 7,120,466 confirmed cases of COVID-19 were reported by 8 June 2020, leading to 406,660 deaths in 215 countries/territories/areas (http://www.worldometers.info/coronavirus/#countries). COVID-19 is reported from most of the countries with the highest total deaths in Italy, Spain, the USA, France, the UK, Iran, China, Germany, Netherlands, and Belgium, in sequence. This virus is highly transmissible, probably through mutations in the viral receptor-binding domain (RBD) that can increase its attachment to the host receptor angiotensin-converting enzyme (ACE2), local transmission by pre-symptomatic people and patients with milder symptoms, and travel of infected passengers to other countries, which could explain the widespread distribution of the disease in these countries (3-5).
The laboratory detection of SARS-CoV-2 in clinical samples of symptomatic patients showed its worldwide explosive spread. However, the underestimation of the cases seems to be seen in most settings, which is mainly due to weakness in laboratory methods and screening of infected cases, especially pre-symptomatic cases or those with milder syndrome (6). A higher fatality risk among cases with laboratory-confirmed tests was reported in China. Based on the last report of the National Institute for Infectious Diseases of Japan, nearly half of the laboratory-confirmed cases were asymptomatic at the time of confirmation, which shows the importance of the selection of appropriate laboratory tests and their impact on the outbreak containment (6).
Based on the WHO definition in its guidance on 27 February 2020 and 18 May 2020, a confirmed COVID-19 case is a person with laboratory confirmation of COVID-19 infection, irrespective of clinical signs and symptoms (7, 8). This definition emphasizes the usage of laboratory tests with good quality, highest sensitivity and specificity, the lowest limit of detection, and lowest false-positive and false-negative results. Preparedness of referral diagnostic laboratories in each country could help governments to contain the outbreak. The tests used by these laboratories should be done with the lowest turn-around-time and highest accuracy. The application of reliable and rapid methods enables clinicians to better manage milder and probable cases, which will be effective in the containment of the outbreak by limiting its local and global transmission. Several diagnostic tests have been developed by different companies, many of which have been rejected by reference laboratories. The diagnostic methods for the primary screening of SARS-COV-2 patients could vary in different countries based on their capacity and resources. The selection of a reliable, rapid, simple, and sensitive method could be helpful in regions with lower resources and will reduce the mismanagement of patients and decreases the number of new cases. This is a challenge due to the urgent need for access to laboratory tests for screening and confirmation of the infection, mainly in symptomatic cases. This systematic review describes and compares the developed diagnostic methods for the detection of SARS-COV-2, based on increasing data on the sequence diversity of this virus and its immunogenicity and pathogenicity.
2. Evidence Acquisition
2.1. Study Identification and Selection of Data
An initial search using keywords “Detection”, “Coronavirus 2019”, “SARS-CoV-2”, and “Sensitivity” identified a total of 526 articles from PubMed and Google Scholar web sites. The exclusion criteria included duplicate studies and research not aiming to develop or validate diagnostic methods for the detection of SARS-CoV-2 in clinical samples. Newly developed or commercial products that did not present approved documents for the report of sensitivity or specificity of the designed tests were also excluded from the study. Based on differences in gold standards and methodologies of the studies, as well as severe biases among them, a qualitative analysis instead of a meta-analysis was done to find the closest findings to reality, and present more accurate and valuable results, their strengths, and weaknesses.
3. Results
3.1. Search Results
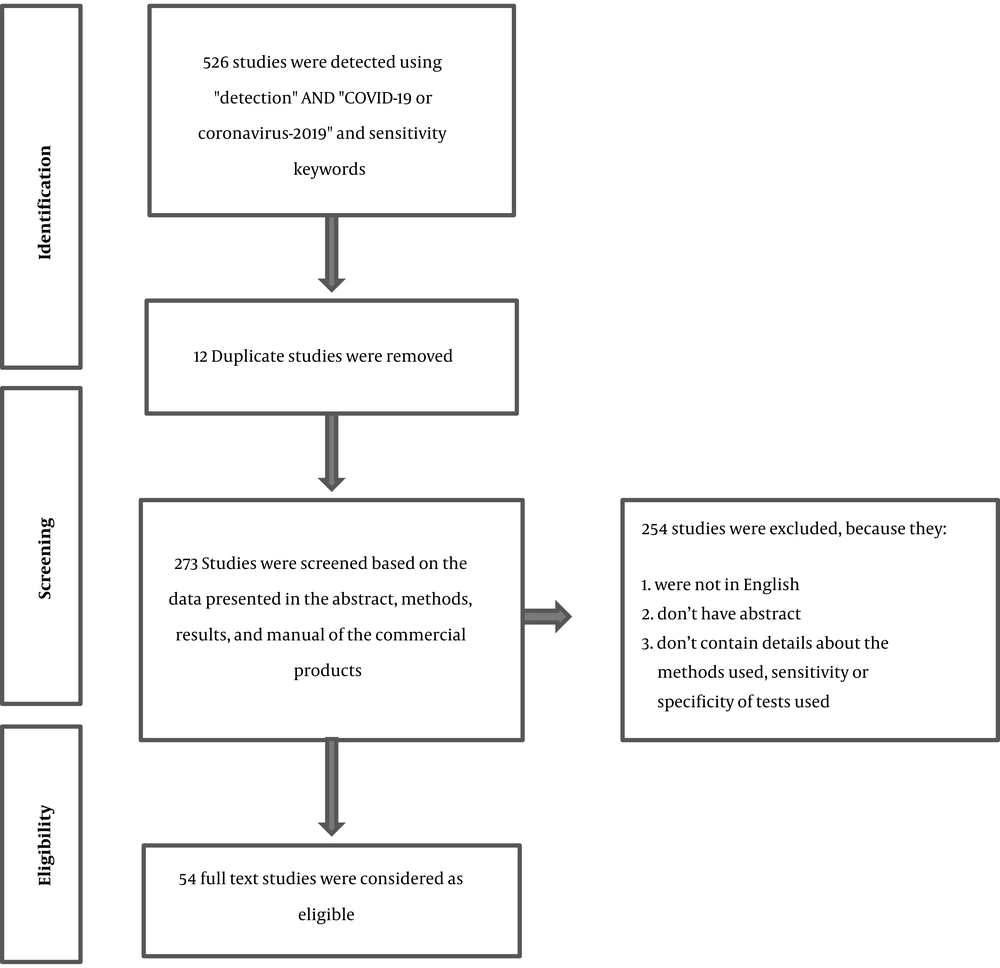
A total of 526 articles were obtained. After removing duplicate and irrelevant results based on the exclusion criteria, 54 studies were selected for the full review (Figure 1 and Table 1). Our searches in general web sites showed that a high number of commercial kits were approved or recorded as in-development for the detection of SARS-CoV-2 (commercial kits for molecular diagnosis and serological tests) by different companies. However, most of them did not provide the sensitivity and specificity of their products. Among the reviewed methods, the applications of serological tests and immunohistochemistry were described in 13 studies, while CRISPR-Cas and target-based amplification methods, including real-time PCR, were presented in 33 studies. The detection limits 1 to 105 copies/µL were reported for the diagnostic methods, irrespective of types of samples used, and disease stages.
| No. | Tests | Type of Sample | Limit of Detection | Sensitivity | Specificity | Reference method | Turn Around | Kit | References |
|---|---|---|---|---|---|---|---|---|---|
| A | Serological Assays | ||||||||
| 1 | Enzyme-linked immunosorbent assay (ELISA) IgG and IgM antibodies | Serum-COVID-19 vs. Serum of healthy people | 55/63, 87.3% | 100% | RT-PCR, WHO guidelines (NAAT) | ND | IgG/IgM antibody ELISA kits were manufactured by Zhu Hai Liv Zon Diagnostics Inc. | Xiang et al. (9) | |
| 2 | COVID IgM/IgG antibodies kit | Blood of confirmed and not-confirmed cases | A Cut-off value of 0.88 and 1.02 for IgM and IgG, respectively | Positive rate increased when a combination of IgM and IgG tests was used | Nucleic acid test | ND | Diagreat company | Jia et al. (10) | |
| 3 | Chemiluminescence | Serum | 86.9% | 99.2% | Total Ab compared to RNA only or IgM only tests | ND | Xia et al. (11) | ||
| ELISA | 94.8% | 100% | |||||||
| Colloidal gold | 96.2% | 95.2% | |||||||
| 4 | Immunohistochemistry sandwich ELISA quantum dots (QD) immunochromatographic technology (synthetic peptides corresponding to different areas of Nucleoprotein (N)) | Blood of immunized mouse with synthetic peptides and lung biopsy of a confirmed patient | 100 ng/mL 10 ng/mL | Not presented | Minimal cross-reaction with SARS and MERS-CoV | ND | ND | In-house | Li et al. (12) |
| 5 | ELISA assay for IgM and IgG antibodies against N protein of SARS-CoV-2 | Serum of confirmed or suspected patients | ND | 83% | 100% | RT-PCR assays and clinical findings | ND | Enzyme-Linked Immunosorbent Assays kit (Lizhu, Zhuhai, China) | Liu et al. (13) |
| 6 | Double-antigens sandwich immunoassay (Ab-ELISA); (receptor-binding domain (RBD) of the spike protein) | Plasma | 89.6% and 100, for days 8-14 and 15-39 after the onset, respectively | 99.10% | RT-PCR | ND | Enzyme linked immunosorbent assay (ELISA) kits by Beijing Wantai Biological Pharmacy Enterprise Co., Ltd., China | Zhao et al. (14) | |
| IgM | 73.3% and 94.3% | 98.60% | |||||||
| IgG | 54.1% and 79.8% | 99.00% | |||||||
| 7 | Point-of-care lateral flow immunoassay (LFIA) test- A rapid IgM‐IgG combined antibody test | Blood, serum, and plasma | 88.66% | 90.63% | RT-PCR and clinical findings | 15 minutes | Jiang et al. (15) | ||
| 8 | In-house and commercial ELISAs (S1-specific IgG and IgA antibodies) | Serum | ND | 100% | Plaque-reduction neutralization test | ND | In-house and commercial kits (EUROIMMUN Medizinische Labordiagnostika AG) | Okba et al. (16) | |
| 9 | ELISA (IgM and IgG antibodies against recombinant nucleocapsid and recombinant spike proteins) | Serum | 80.4 - 82.2% | 100% | RT-PCR | Lizhu, Zhuhai, China and Hotgen, Beijing, China | Liu et al. (17) | ||
| 10 | Colloidal gold-immunochromatographic assay (GICA) | Serum-COVID-19 vs. Serum of healthy people | 75/91, 82.4% | 100% | RT-PCR | IgG/IgM antibody GICA kits (lot number 2001010220), manufactured by Zhu Hai Liv Zon Diagnostics Inc, China | Xiang et al. (9) | ||
| 11 | Peptide-based luminescent immunoassay (magnetic chemiluminescence enzyme immunoassay, MCLIA) to detect IgG and IgM (synthesized peptides from the orf1a/b, spike (S), and nucleocapsid (N)) | Serum of confirmed cases with RT-PCR | Mean chemiluminescence (CL) values for IgG and IgM as 0.152 ± 0.109 and 0.151 ± 0.107, respectively | 71.4%, 57.2%, and 81.5% for IgG, IgM, and total antibody, respectively | 100% | RT-PCR | ND | Cai et al. (18) | |
| 12 | Fluorescence immunochromatographic assay (Nucleocapside protein) | Nasopharyngeal swab sample and urine | 68% | 100% | RT-PCR | within 10 minutes | Diao et al. (19) | ||
| 13 | Colloidal gold-based immunochromatographic strip (IgG and IgM) | Serum, plasma, and whole blood | 68.6% in total | RT-PCR and clinical findings | 15 min | Zhuhai Livzon Diagnositic Inc. | Pan et al. (20) | ||
| 92.9% in the intermediate stage (8 - 14 days of onset) | |||||||||
| 96.8% in the late stage (≥ 15 days of onset) | |||||||||
| B | Molecular Assays | ||||||||
| 14 | qRT-PCR (ORF1ab and nucleocapsid protein (N)) | Throat swab samples, sputum samples, and alveolar lavage fluid samples of suspected cases | 50 copies/μl to 105 copies/μl | 42/81 (51.9%) | RT-PCR | 2019-nCoV nucleic acid detection kit according to the manufacturer’s protocol (Shanghai ZJ Bio-Tech Co Ltd) | Xiang et al. (9) | ||
| 15 | qRT-PCR (N and RdRP) | N has 7 - 43 fold greater sensitivity than RdRP; ~10 fold increased sensitivity | RT-PCR | PCLMDTM nCoV one step RT-PCR Kit from PCL Inc, Korea, Cat # PCLMD2019001-50 | Kim et al. (21) | ||||
| 16 | Automated RT-qPCR (N1, E, and RdRP) | 21, 141, and 350 copies/reaction for N1, E, and RdRP, respectively | N1 and RdRP showed the highest analytical sensitivity | RT-PCR | Barra et al. (22) | ||||
| 17 | Droplet digital PCR (ddPCR) (ORF1ab and N) | Pharyngeal swab samples from suspected patients | 0.109 copies/µL and 0.021 copies/ µL for ORF1ab and N primers/probe sets | 500 times higher than qRT-PCR | RT-PCR | QX200 Droplet Digital PCR System using supermix for probe (Bio-Rad) | Suo et al. (23) | ||
| 18 | qRT-PCR vs. CT scan | Throat and nasal swabs | Between 45 and 60% vs. 97% | RT-PCR | Al-Tawfiq et al. (24) | ||||
| 19 | qRT-PCR assay (RdRp/Orf1 and N region) | 15 copies/reaction; ORF1ab (China), 2019-nCoV-N2, N3 (USA), and NIID-2019-nCOV-N (Japan) sets | ORF1ab: ORF1ab (China) set might be most sensitive than other sets; N: 2019- nCoV-N2, N3 (USA), and NIID-2019-nCOV_N (Japan) sets may be recommended for the sensitive qRT-PCR assay of N region | RT-PCR | Jung et al. (25) | ||||
| 20 | qRT-PCR | Nasopharyngeal swabs or sputum | 10 copies/reaction | RealStar SARS-CoV-2 kit seemed to be more sensitive | RT-PCR | 1:50 - 2:15 hrs | QuantiTect Virus + Rox Vial kit (QIAGEN); SuperScript III One-step RT-PCR System with Platinum TaqDNA Polymerase (Invitrogen); RealStar SARS-CoV-2 RT-PCR kit 1.0 (Altona) | Konrad et al. (26) | |
| 21 | qRT-PCR ((RdRp)/helicase (Hel), spike (S), and nucleocapsid (N) genes) | Respiratory and non-respiratory samples | 11.2 RNA copies/reaction | COVID-19 -RdRp/Hel showed higher sensitivity vs. RdRp -P2 assay | More specific vs. RdRp-P2 assay | RT-PCR | Chan et al. (27) | ||
| 22 | Multiplex-PCR-based method and multiplex-PCR-based metagenomic method | 1.15 copies | ND | ND | RT-PCR | Li et al. (28) | |||
| 23 | Digital PCR (dPCR) (ORF 1ab, N and E genes) | Pharyngeal swab | 2 copies/reaction | Higher than RT-qPCR | 100% | RT-qPCR | Dong et al. (29) | ||
| 24 | RT-LAMP (iLACO) | ND | 10 copies of ORF1ab/reaction | 97.6% (42/43) vs. RT-PCR | RT-PCR | Yu et al. (30) | |||
| 25 | RT-LAMP coupled with nanoparticles-based biosensor (NBS) assay (RT-LAMP-NBS) (orf1a/b and N) | Oropharynx swab | 12 copies/reactions | 100% | 100% | RT-PCR | 1 h | Zhu et al. (31) | |
| 26 | Nanopore target sequencing (NTS) of 11 related gene fragments of orf1ab | Nasopharyngeal swab | 10 copies/mL | More sensitive than qRT-PCR | RT-PCR | 6 - 10 h | Wang et al. (32) | ||
| 27 | Bait capture platform for culture- and amplification-free next-generation sequencing of SARS-CoV-2 | In silico | ND | ND | ND | ND | ND | ND | Nasir et al. (33) |
| 28 | Genome Detective Coronavirus Typing Tool | 100% | 100% | A reference dataset of previously published coronavirus whole-genome sequences | Cleemput et al. (34) | ||||
| 29 | SENsitive Splint based one-step isothermal RNA detection (SENSR) (RdRp) | RNA from clinical samples | 0.1 aM, 6 molecules per 100 µL reaction | ND | ND | ND | 30 min | Woo et al. (35) | |
| 30 | Combination of RT-PCR and CT, Two targets: RdRp+, E or N+ | Pharyngeal and nasal swabs and stool | 91.90% | 66.8% RT-PCR is superior to CT in diagnosing mild infections | RT-PCR, WHO guideline | ND | ND | Jiang et al. (36) | |
| 31 | Two monoplex real-time RT-PCR assays targeting ORF1b and N gene regions | Sputum and cell culture supernatant | ≥ 10 copies of chimeric plasmid | N gene assay is about 10 times more sensitive than the ORF-1b gene assay | RT-PCR | In-house method | Chu et al. (37) | ||
| 32 | RT-PCR (E and RdRp) | Respiratory samples | 5.2 and 3.8 copies per reaction at 95% detection probability, respectively | 100% | RT-PCR | World Health Organization | Corman et al. (38) | ||
| 33 | Real-time RT-PCR (ORF1ab, nucleocapsid protein-NP) | Nasal, pharyngeal swabs, bronchoalveolar lavage fluid and sputum | Not presented | Not presented | RT-PCR | Shanghai Huirui Biotechnology Co., Ltd. | Liu et al. (39) | ||
| 34 | Real-time reverse-transcription PCR (rtPCR); RdRP, S, E, N | Pharyngeal swab | 1 - 10 virus particles | High | Not presented | RT-PCR | less than 4 hours | Won et al. (40) | |
| 35 | CRISPR-Cas12 (RdRp, ORF1b and ORF1ab genes) | Saliva | 10 copies/µL | ND | Lucia et al. (41) | ||||
| 36 | CRISPR-based DETECTR Lateral Flow Assay; CRISPR-Cas12 based (E and N genes + human RNase P gene as a control) | Respiratory swab | 10 copy/µL reaction (in vitro); Decreased to 500 and 1,500 copies/µL in raw samples carried in transport media and PBS | 90% | 100% | RT-PCR | 30 min (RNA)- 45 min (Sample to results) | Broughton et al. (42) | |
| 37 | CRISPR-Cas13 detection system (Orf1ab) | 10 copies/µL | Not presented | Not presented | ND | Metsky et a. (43) | |||
| 38 | CRISPR/Cas13-mediated enzymatic signal amplification (Orf1ab) | Respiratory samples | 1 - 7.5 copies | 100% | 100% | Metagenomic next-generation sequencing (m-NGS) | 40 min | Hou et al. (44) | |
| 39 | All-in-One-dual (AIOD)-CRISPR (CRISPR-Cas12a) | In vitro | 1.2 copies | ND | ND | RT-PCR | 40 min | Ding et al. (45) | |
| 40 | LAMP (ORF1a-A, GeneN-A) | Respiratory specimens (swabs) | 4.8 copies/µL | In agreement with the real-time detection | RT-PCR | Zhang et al. (46) | |||
| 41 | RT-LAMP (ORF1ab gene, E gene and N gene) | Throat and nasal swabs | 1000 copies/mL (5 copies) | Similar to RT-PCR. The ORF1ab gene was very specific and the N gene was very sensitive | 99% | RT-PCR | In-house RT-LAMP vs RT-PCR (Jiangsu Bioperfectus Technologies Co., Ltd.) | Yang et al. (47) | |
| 42 | Single-step reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) | Spiked molecular grade water and sputum samples | 500 copies/mL | 91.4% | 99.5% | qRT-PCR | < 30 min | Jiang et al. (48) | |
| 43 | RT-LAMP (Nsp3, S, N) | In vitro | 100 copies | ND | 100% | ND | 30 min | Park et al. (49) | |
| 44 | RT-ERA: Isothermal exponential amplification methods via reverse transcription plus subsequent enzymatic recombinase amplification (ERA); (N-gene); nfo affinity probe plus LF strips | In vitro | 0.32 aM (i.e. 0.32 × 10 - 18 M) of RNA or 0.2 copy/µL | ND | ND | ND | 30 min | Xia et al. (50) | |
| 45 | closed-tube Penn-RAMP, a two-stage; isothermal dsDNA amplification method | ND | Greater than LAMP or RT-PCR alone | ND | > 1 h | Yang et al. (51) | |||
| 46 | Double-quencher probes (YCH assay) (N1 and N2) | In vitro | > 10 copies of DNA positive control | Greater than US CDC and CDC) in the USA and the National Institute of Infectious Diseases (NIID) in Japan | RT-PCR | Hirotsu et al. (52) | |||
aNot reported or detected; RT-PCR: Reference real-time PCR method reported by the WHO or other international organizations as described in each article.
3.2. Diagnostic Methods for SARS-CoV-2
3.2.1. Serology
Research studies on infected patients with SARS-CoV-2 showed the immunogenic nature of the virus in different populations. Increases in IgM, IgA, and IgG antibody titers were confirmed by the newly developed serological tests, as these assays could provide valuable data about the epidemiology of the virus. Serological assays could be used for the detection of infected symptomatic and even asymptomatic cases at the end of the disease course and thereafter. Although no universal serological tests are now available for the detection of active infection, the application of such tests can be useful for the detection of prior infection among symptomatic and asymptomatic people in different populations. The drawback of SARS-CoV-2 antibody tests for the diagnosis of COVID-19 infection was also presented in the latest update of the WHO guideline (8). In spite of this shortcoming, ELISA and other antibody-based immunoassay techniques, such as immune colloidal gold detection technique, immunohistochemistry, sandwich ELISA quantum dots (QD) Immunochromatographic technology, point-of-care lateral flow immunoassay, and peptide-based luminescent immunoassay, are among the developed immunoassay methods for the direct or indirect detection of SARS-CoV-2 in human sera, plasma, or whole blood samples (9, 10, 12, 13, 15, 18, 53). Currently, more than 170 immunoassay methods are available, only one of which, named “qSARS-CoV-2 IgG/IgM Rapid test” by Cellex Inc., has been approved by the Food and Drug Administration (FDA). Rapid chromatographic immunoassay kits, such as COVID-19 Ag GICA kit, rapid IgM-IgG combined antibody test, and qSARS-CoV-2 IgG/IgM Rapid Test, can detect SARS-CoV-2 antigen or antibody in nasopharyngeal swabs or blood samples and their results are accessible within 10 - 30 min. The point-of-care lateral flow immunoassay (LFIA) test, a rapid IgM-IgG combined antibody test, showed the lowest turn-around time (15 min); however, its specificity was as high as 90.63%. While there are some reports about the accuracy of these rapid immunoassay kits, there are no publications to refer to the obtained results.
The sensitivity of serological methods mainly depends on the number of days post-infection; it can reach 100% after 15 - 39 days of disease onset. The sensitivity of serological methods was greater than 90% in most of the assays. However, a cross-reaction with SARS and MERS-COV was reported in an in vitro immunohistochemistry sandwich ELISA quantum dots (QD) Immunochromatographic technology assay (12). A combination of IgG and IgM antibody tests against SARS-CoV-2 showed an increased positivity rate compared to single IgG or IgM tests or molecular methods (10). In another study, the combination of serological tests (total antibody) and molecular methods (RT-PCR) showed a 12% higher sensitivity rate, suggesting ELISA + RT-PCR a better diagnostic indicator, even for the early diagnosis of COVID-19 infection (within seven days after disease onset). These results show that serological assays could play a role as a supplement to RNA detection methods to improve diagnostic sensitivity and specificity (11).
3.2.2. Molecular Methods
Target specific detection, amplification, or sequencing of genetic materials of SARS-CoV-2 in clinical samples could be done by different molecular methods. These methods can detect and/or amplify the target sequences using degenerative primers, multiple primer sets, or a single set of primers. At the time of writing this article, more than 202 molecular assays had been developed for the detection of SARS-CoV-2, 168 of which were commercialized (https://www.finddx.org/covid-19/pipeline/) and 60 products were CE-IVD-certified. The validated products included Caspr Biotech Phantom 1.0 Dx (near-point-of-car [POC]), Coyote Bioscience 2019-nCoV Prep Free QPCR Assay (near-POC), Fluxergy LLC Fluxergy Sample-to-Answer SARS-CoV-2 RT-PCR, General Biologicals Corporation GB SARS-CoV-2 Real-Time RT-PCR, Integrated Nano-Technologies Palladium, and STILLA COVID-19 Multiplex Crystal Digital PCR detection kit. Other molecular methods that their applications for SARS-CoV-2 were studied include the electrical detection of SARS-CoV-2 nucleocapsid protein using nanosensors and aptamer, NGS-based target capture for SARS-CoV-2 detection and screening, loop-mediated isothermal amplification (LAMP), Colorimetric and Isothermal Detection Kit, and CRISPR (clustered regularly interspaced short palindromic repeats)-Cas systems. As the RT-PCR methods, the WHO (https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance) and the US Centers for Disease Control and Prevention (https://www.cdc.gov/coronavirus/2019-ncov/lab/index.html) developed RT-PCR Diagnostic Panels for the universal detection of SARS-like betacoronaviruses and the specific detection of SARS-CoV-2. Other reference laboratories also proposed alternative RT-PCR assays that generally target conserved gene loci of the virus in RNA extracts of clinical samples. These assays initially do a screening and therefore need to complete two or three sets of reactions to confirm the results. The integration of these tests into a multiplex real-time PCR panel has improved their application for screening of suspected patients during the pandemic in most countries.
In a comparative study on metagenomic-NGS (mNGS), RT-PCR, and CRISPR, in which mNGS was used as the reference method, PCR- and CRISPR-nCoV both presented a specificity of 100%. Besides, RT-PCR could detect the virus in 90.4% of the positive cases, which was explained due to the lower titer of the virus in some of the studied samples. However, CRISPR showed greater sensitivity (100%). In this study, higher sensitivity and lower turn-around-time of the CRISPR system (RT-PCR took about 1.5 hours, mNGS approximately 20 hours, and CRISPR around 40 min) and its usefulness in under-resourced settings where thermal cyclers are not accessible suggested the superiority of this technology over other methods for the detection of SARS-CoV-2 in clinical samples (44). In a recent review of the detection of human coronavirus infections, 17 diagnostic methods were summarized as cleared Chinese kits for the laboratory diagnosis of SARS-CoV-2 infection. The methods used by these products were included fluorescence RT-PCR, NGS, immune colloidal gold technique, isothermal amplification and microarray, magnetic particle chemiluminescence, and chemiluminescence immunoassay (54). However, the sensitivity and specificity of the cleared diagnostic kits were not presented clearly.
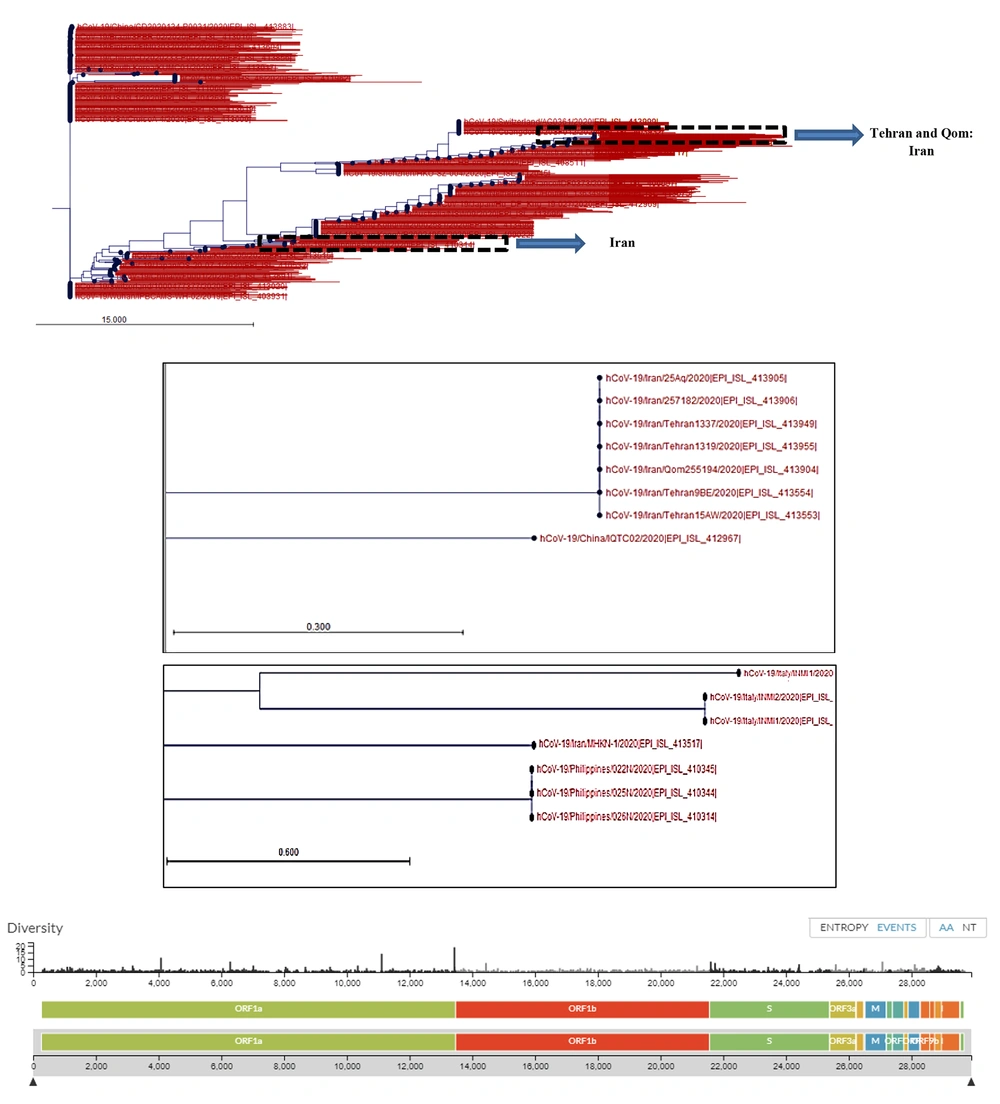
The PCR and real-time PCR methods are among the common test methods developed by different companies and national or international health authorities. All of these methods target ORF1AB, spike (S), nucleocapsid (N), envelope (E), and RNA-dependent RNA polymerase (RdRP) genes. The assays were tested on RNA extracts of saliva, nasal, and pharyngeal swabs, bronchoalveolar lavage fluid, sputum, and stool samples. The detection limits of the molecular assays reviewed in our study ranged between 0.109 copies/µL and 105 copy/µL and their sensitivity was between 45% to 97.6%. The highest sensitivity was reported for the RT-LAMP (iLACO) method used for the amplification of the ORF1AB gene when compared to the RT-PCR method (97.6%). Among the targeted genes, ORF1AB showed the highest specificity and the N gene showed the highest sensitivity for the detection of SARS-CoV-2 RNA in clinical samples using different molecular methods. The analysis of sequence diversity among different loci of the SARS-CoV-2 genome in strains recovered from different countries showed a nucleotide substitution rate of 8 × 104 substitutions per site per year (Figure 2). These substitutions were also depicted at gene loci that were mainly targeted for the amplification and diagnosis of the infection in RNA extracts of clinical samples using molecular methods reviewed in this article. This diversity was not limited to different geographic locations but was reported among isolates in each country (Figure 2).
Phylogenetic tree of SARS-CoV-2 strains recovered from different countries around the world. Phylogenetic tree was drawn using CLC Genomic Workbench 11.0.1 based on the released sequences of SARS-CoV-2 from different countries, including Iran. The sequence of Wuhan-Hu-1/2019 was used as the reference. The Iranian strains were clustered at two distinct locations, which were originally related to one strain from China (seven Iranian strains, Upper dashed box) and one strain isolated from Philippine (Lower dashed box). Nucleotide diversity of SARS-CoV-2 isolates at nucleotide levels was provided from GISAID-Global Initiative on Sharing All Influenza Data-database based on the shared data updated on 2020-03-20.
In a study of isothermal amplification methods, researchers combined isothermal amplification with CRISPR-Cas12 DETECTR technology to develop a rapid detection method for SARS-CoV-2 (42). In this assay, RNA extracts of nasopharyngeal or oropharyngeal swab samples were used for the detection of N and E genes as targets. The assay was comparable in accuracy to the qRT-PCR method approved by the WHO and US CDC. Low cost, minimum requirement of laboratory equipment, and short turn-around-time (30 - 45 min) were proposed as its advantages. In a study by Song et al., a closed-tube Penn-RAMP, a two-stage isothermal dsDNA amplification method, was developed for the detection of SARS-CoV-2. The assay utilized both recombinase polymerase amplification (RPA) and LAMP techniques in a single tube. This RNA-based point-of-care (POC) technology combines both a paper-based POC diagnostic device and LAMP, which can be integrated with a smartphone application.
3.2.3. Proteomic Assays
Among the other proposed detection methods for SARS-CoV-2, shotgun proteomics were proposed in an in silico study for screening of this virus in proteolytically digested body fluids (55). In this method, digested viral peptides are screened by liquid chromatography-mass spectrometry (LCMS systems). However, no in vitro test on human clinical samples was done for the validation of the proposed method. The application of the fluorescence immunochromatographic assay that uses microbial antigens for diagnosis was proposed in a study of 239 participants suspected to COVID-19 infection. The assay could detect the N protein in either swab or urine samples of patients in the early phase (3 - < 5 days after clinical onset) of the disease occurrence (19). In this study, the existence of the N antigen in nasopharyngeal swabs was detected in all participants that had positive RT-PCR results (ORF1ab and N). Although the performance of the assay on the nasopharyngeal swab samples showed a sensitivity of 68% compared to RT-PCR, it showed a capacity for the detection of the virus in ~74% of urine samples among patients with positive nasopharyngeal antigen tests.
4. Discussion
In this review, due to the numerous biases in studies and the inconsistency of studies in terms of gold standard methods, it was not possible to perform meta-analyses on their results. Thus, it is necessary to use qualitative analyses to obtain some answers about the diagnostic accuracy of tests. The researchers of this study admit that because of the bias in the studies, it is not possible to draw definite conclusions from them, but based on the analysis of the effect of each type of bias on the results, the effect of compensatory measures used by each study in this field, and comparing the results of different studies, it can be seen which of the results of the study is closer to reality. We also use logical inferences in qualitative analysis to judge the results of studies.
To analyze the validity of the results of the studies used in this review, the following factors were considered:
4.1. The Sampling Method and Correct Diagnostic Study Protocol
In most of the studies, infected and healthy samples were selected separately; that is, patient samples were selected from approved patients, and healthy samples were selected from individuals who did not have any specific viral disease. Known as spectrum bias, this bias is one of the most important factors that disrupt the results of sensitivity and specificity estimates. In studies that have this type of bias, both indicators are usually shown to be greater than the actual value. Studies by Xiang et al. (9), Zhao et al. (14), Li et al. (12), Yu et al. (30), Zhu et al. (31), Cleemput et al. (34), Broughton et al. (42), and Jiang et al. (48) had this type of bias. Studies by Liu et al. (17) and Cai et al. (18) partially encountered this problem although they tried to compensate to some extent by adding people with other diseases. Studies by Diao et al. (19), Jiang et al. (36), and Yang et al. (47) did not have this type of bias, but for reasons that we will discuss below, their results were not conclusive.
4.2. How to Choose the Gold Standard and the Final Confirmation of Virus Infection?
In most of the studies, the final confirmation of contamination was based on the two guidelines by WHO and a guideline from China; given that the final confirmation in these two guidelines was based on NAAT and RT-PCR, respectively, the sensitivity and specificity of tests that perform the same function as NAAT cannot be judged correctly. In other words, the sensitivity and specificity of these cases were falsely higher than the actual value. This problem can be seen even in studies that have not had spectrum bias.
4.3. Indirect Conclusions From the Data
Since the direct conclusions are erroneous for the above reasons, we tried to obtain an estimate of the correctness of the reported indicators based on indirect conclusions from the data. The findings by Liu et al. (17) and Pan et al. (20) are significant in this regard. In the studies by Liu et al. (17), out of 238 patients who were clinically highly suspicious, 85 cases were reported to be RT-PCR-negative. In this study, the ratio of positive antibodies in both approved and unapproved groups and their clinical status were similar. In the Pan et al. (20) study, there were 95 RT-PCR-confirmed samples and 39 RT-PCR-negative samples, which were clinically confirmed. In this study, the ratio of positive antibodies in both approved and unapproved groups was similar, and the results were similar to the findings in the Liu et al. study (about 80% of their antibody tests were positive). These results indicate that the antibody and RT-PCR tests are epidemiologically independent of each other.
The study’s authors conclude that patients who are highly suspicious in terms of clinical symptoms can be considered definitive. Logically, this conclusion is correct because when an independent test (antibody test) in both PCR-positive and PCR-negative independent populations, is positive by 80%, it can be proven by using a mathematical argument that all people in the PCR-negative population are sick. Otherwise, the antibody test will be 80% false positive, and since this is not possible, the only assumption is that both groups are sick. Based on the above arguments, it can be concluded that:
Clinically highly suspicious cases are similar to those in which RT-PCR is positive, and both groups can be considered definite patients (the author has done the same).
A 64% sensitivity for RT-PCR is close to reality.
Because antibody tests and RT-PCR are epidemiologically independent of each other, they can complement each other.
In a study by Pan et al., 70 people were examined as patients other than COVID-19, and the results of antibody testing in four of them showed a false positive. This finding confirms that a 100% specificity is incorrect for the ELISA test and it is due to the effect of spectrum bias. Based on the above findings, it can be said that the sensitivity of the tests whose gold standard was RT-PCR was much lower than the reported value (considering that the sensitivity of RT-PCR is 64%). A study by Jiang et al. (36), which examined the sensitivity and specificity of RT-PCR based on the WHO guidelines and could be considered conclusive in terms of the sampling method, reported a 78.2% sensitivity for RT-PCR. However, considering that the NAAT method is the gold standard in the WHO guideline, and because of its similarity to RT-PCR in targeting sites, this study may have undergone an overestimation of RT-PCR sensitivity and specificity.
Another point in the findings of the above studies by Liu et al. (13) and pan et al. is that the method chosen by these two studies to find definitive patients minimized bias incorporation, while the result of Ren et al. study was affected by this bias.
Based on the foregoing discussion, it can be said that the sensitivity results cannot be cited in any of the reviewed studies, except for Liu et al. (17) and Pan et al. (20) studies. The results of specificity in any of the studies, even two studies by Liu et al. (17) and Pan et al. (20) cannot be cited due to limitations in the selection of the healthy group.
5. Conclusions
The RT-PCR test alone cannot act as a gold standard method. Since the results of antibody tests and other methods are independent of RT-PCR, they can be used as supplementary diagnostic tests. Although clinical findings are invaluable for the direct diagnosis of the infection, in many cases, they can be considered more valuable than serological tests. Based on the findings from the analysis of the study results, it is recommended that future COVID-19 diagnostic studies use combined gold standards with full follow-up, because trusting on a test as the gold standard leads to bias in measurements. Clinical prediction rule studies can be helpful to find an effective diagnostic pathway.