1. Context
In December 2019, several patients suffering from pneumonia were admitted to a hospital in Wuhan, China. Subsequently, they were diagnosed with a new coronavirus disease, i.e., COVID-19, which had spread to more than 170 countries all around the world. By the time this draft was being revised (May 2021), over 150 million cases and three million deaths were confirmed globally by the World Health Organization (WHO) (approximately 2.1% case fatality rate [CFR]) (1, 2). The CFR of COVID-19 is considerably lower than that of severe acute respiratory syndrome (SARS) (approximately 9.5%) and much less than Middle East respiratory syndrome (MERS) (around 35%) (3). To compare the similarity of the three viruses, next-generation sequencing (NGS) analysis of the specimen containing SARS-CoV-2 has shown 79 and 50% sequence homology to SARS-CoV and MERS-CoV, respectively (4). The difference in sequence homology is partially associated with how these viruses enter the host cells. With this in mind, both SARS-CoV and SARS-CoV-2 exploit angiotensin-converting enzyme 2 (ACE-2) receptor, while MERS-CoV employs dipeptidyl peptidase 4 (DPP4, also known as CD26) (5-7). The binding of the spike glycoproteins of SARS-CoV-2 (2SP) to ACE-2 enables the process of cell entry (8, 9).
As most viruses, including coronaviruses, are generally dependent on the host cells to replicate and further the progression, the cytokine storm is being exploited to destroy the cells and the occurrence of multi-organ failure. The expression balance of transcription factors associated with the pro-inflammatory cytokines in non-infected cells gets lost when SARS-CoV-2 enters the cells. Subsequently, it utilizes the host cells’ enzymes, and the cells undergo an imbalanced situation, leading to organ failure (10, 11).
Several years since the outbreak of SARS and MERS, no particular cure has yet been found (12, 13). That is to say, the absence of a promising drug to eliminate COVID-19 is profoundly felt. This review investigated the potential targets for the treatment of SARS-CoV-2, including ACE-2 and other key molecules contributing to SARS-CoV-2 infection. The importance of these molecules in COVID-19 was somewhat clarified in previously published articles. But the focus on the employment of novel and repurposed drugs was hampered as a result of the vague aspects to the signalling process of these elements. Several potential targets are introduced to be further analyzed for the treatment of COVID-19.
2. Evidence Acquisition
In this review, a literature search was implemented in English from 6 October 2019 to 2 May 2021. The search was performed in electronic databases, including PubMed, Scopus, and Google Scholar. In the present review, according to the medical subject heading (MeSH) criteria, the following keywords were used: "SARS-CoV-2", "COVID-19", "ACE-2", "Therapeutic Targets", "Acute Respiratory Distress Syndrome", and "Cytokine Storm."
3. Results
3.1. SARS-CoV-2 Structure and Pathogenicity
SARS-CoV-2 and SARS-CoV belong to the lineage B of the betacoronavirus genus, while MERS-CoV is from the lineage C of the same genus (14). The genome of beta coronaviruses is about 30 kb, which generally contains six to 10 genes. The first gene is associated with transcription and duplication, and the other remaining genes code for structural proteins (15). Spike protein is a major structural protein of SARS-CoV-2, along with envelope (E), nucleocapsid (N), and membrane (M) proteins. There are other proteins, including around 16 nonstructural and five to eight accessory proteins (16). Before the viral assembly, most of the viral proteins, specifically N proteins, are in the endoplasmic reticulum-Golgi region. Since there is a connection between this complex and nucleic acid content, it is believed that N protein is associated with the viral replication cycle. M protein forms the structure of the virus by acting as a platform for stabilizing the complex when other proteins are bound to it. The smallest structural protein, i.e., E protein, is responsible for developing newly replicated viruses (17).
It was suggested that the process of viral entry through ACE-2 receptor takes place by two various pathways. The distinction of these two pathways is defined by the process of 2SP priming, either with transmembrane serine proteases or endosomal cysteine proteases (18).
The pathogenicity of SARS-CoV-2 mostly relies heavily on the activity of 2SP as the initiator of binding to the host cells. Its role is crucial for the virus, and therefore, it is considered as the main target for inhibition of SARS-CoV-2 pathogenesis (19).
3.2. Host Cell Receptors and Associated Proteins
Several receptors have been thought to be participating in the process of SARS-CoV-2 cell entry. The entangled host cell receptor proteins could serve as potential drug targets.
3.2.1. The Proteins Involved in SARS-CoV-2 Cell Entry via ACE-2 Receptor
In coronaviruses, S glycoprotein is mainly involved in infecting the host cells. The variation of S glycoprotein among different species of the Betacoronavirus determine the pathogenicity of different members (20). Since SARS-CoV-2 cell entry depends on the binding of 2SP to ACE-2 receptors, the availability and number of ACE-2 receptors are the key factors for furthering the process of infection in alveolar cells (21). ACE-2 receptor distribution in other organs, including the heart, kidney, and intestine, may justify the multi-organ dysfunction in severe COVID-19 cases (22).
The 2SP has two subunits, S1 and S2. The binding of the former to the ACE-2 cell receptor is an essential step for SARS-CoV-2 cell entry. Subsequently, the fusion takes place by the activity of the S2 subunit (19). In terms of domain numbers, the structure of the S1 subunit is analogous to that of S2 (S1 trimer is located on top of S2 trimer). Moreover, each of its domains has its receptor-binding domain (RBD), which contains the receptor-binding motifs (RBM) that primarily account for docking to the cell receptors. Previously, two relatively stable conformations acquired by the S1 domain B in 2SP were detected by cryoelectron microscopy (cryo-EM). Either one domain has open RBD, and the other two are in a closed state, or all three RBDs are enclosed, resulting in evasion of the binding site from the immune system. This finding somewhat clarified the vague aspect of the spike RBD inhibitors not being entirely effectual (23).
To that end, cysteine and serine proteases (Cathepsin B and L (CatB/L) and TMPRSS2, respectively) prime S protein by cleaving different S protein sites (Figure 1) (24). It also has been declared that in comparison with other coronaviruses, the availability of a leading proline residue influences the cleavage loop exposure in a way that the turn created by the proline enhances the activity of the proteases (25).
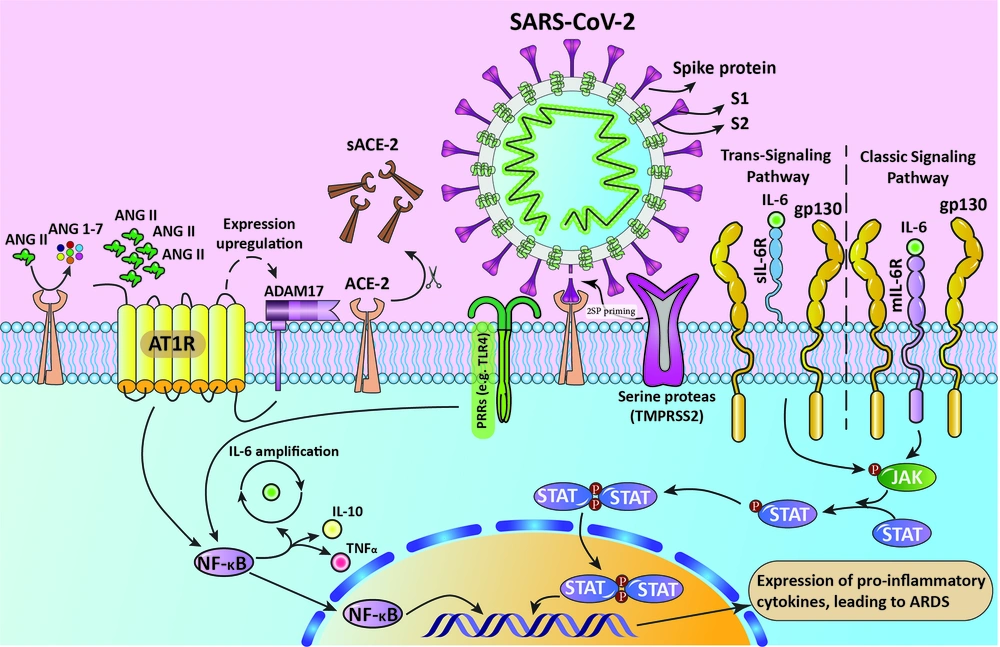
The process of inducing ARDS by 2SP. This figure illustrates that in the presence of SARS-CoV-2, its spike glycoprotein binds to the active site of the ACE-2 receptor and prevents the binding of AngII. Also, ACE-2 is cleaved as a process of AT1R-mediated ADAM17 upregulation, resulting in the accumulation of AngII in serum, which in turn activates the NF-κB pathway. SARS-CoV2 also activates IL-6 production via the NF-κB pathway. IL-6 activates JAK/STAT pathway via transmembrane and soluble IL6R. The activated STAT dimers act as transcription activators and induce the expression of pro-inflammatory cytokines. The process ends in ARDS.
Inhibition of TMPRSS2 and Cat B/L molecules is associated with hampering SARS-CoV-2 cell entry by inhibiting viral fusion (22, 26). It was shown that, in TMPRSS2-negative, ACE-2 expressing cells, while Cat B and L are blocked, SARS-CoV-2 cell entry is completely disrupted. However, in TMPRSS2 positive cells, the inhibition happens with lower efficiency. Since the expression of TMPRSS2 alone is enough for infectivity of SARS-CoV-2, protease activity of CatB/L is dispensable for SARS-CoV-2 infection (19, 27). Hence, available drugs that inhibit the activity of TMPRSS2 could be used as potential therapeutic options. In an in-silico study, two drugs (Rubitecan and Loprazolam) were found to act against TMPRSS2 by structure-based virtual screening. Furthermore, the value of binding energy was measured for these two drugs; the results suggested that Rubitecan is more stable than Loprazolam (binding free energy was calculated using the CaFE tool) (28).
Because the sequence homology of SARS-CoV and SARS-CoV-2 is slightly more than 70% and both use ACE2 for cell entry, the mechanism by which they enter the cells is more or less the same. However, investigating the spike protein of SARS-CoV led to the discovery of leading structural differences that affect the infectivity of SARS-CoV-2. By homology modeling, it was shown that in SARS-CoV-2, the interaction of ACE-2 and four specific residues in the receptor-binding domain (located in the S1 subunit) of 2SP is eliminated compared to that of SARS-CoV S. Instead, four other residues establish new interactions. What is more, a salt bridge existing between the SARS-CoV RBD and ACE-2 is substituted with a new salt bridge in SARS-CoV-2 (29). In another study, it was indicated that considering the complex structure of the SARS-CoV-2 spike C terminal domain, most of the binding sites overlay with SARS-CoV (30). In other studies on SARS-CoV RBD, it is suggested that RBD consists of a core and a receptor-binding motif (RBM) and the S protein interacts with ACE-2 via the RBM. There are two hotspots in ACE-2 that are crucial for their interaction. It was shown that several mutations in the RBM enclose these hotspots, which may justify the difference in the infectivity of SARS-CoV to that of SARS-CoV-2 (31, 32).
3.2.2. ACE-2 and IL-6 Receptors: Impact on the Cytokine Storm in the Opposite Direction
Angiotensin 2 (AngII) is a cytokine accountable for contraction in blood vessels and inflammation by activating intermediate molecules using angiotensin receptor type I (AT1R). Normally, ACE-2 inactivates AngII, but the occupancy of ACE-2 receptors by S proteins results in escalation in the AngII level in serum. Moreover, AT1R is associated with the expression of ADAM17 (A Disintegrin and Metalloproteases 17) protein, which acts by cleaving ACE-2 receptors and solubilizing them, resulting in inhibition of ACE-2 mediated AngII degradation (33, 34). Thereupon, the AT1R-AngII axis activates NF-κB via phosphorylation of its P65 subunit (35).
In another axis, SARS-CoV-2 itself activates NF-κB with the help of pattern recognition receptors (PRRs), leading to an increasing number of IL-6, TNFα, and IL-10 (36). IL-6 is an inflammation mediator which acts by prompting JAK/STAT pathway. The increased production of IL-6 enhances the binding of IL-6 to its receptors (IL-6R). IL-6 acts through binding to two different receptors, transmembrane receptor (mIL-6R) and soluble receptor (sIL-6R), which is the cleaved form of mIL-6R dissociated by ADAM17 (37). Provided that if it binds to mIL-6R or sIL-6R, classic signaling and trans-signaling pathways activate, respectively. The types of cells that express mIL-6R are limited, and due to the availability of soluble IL6 receptors everywhere in the body, the trans-signaling pathway is considered to be associated with cytokine storm, especially in acute respiratory distress syndrome (ARDS) (Figure 1) (37, 38).
As IL-6 binds to the soluble receptor, the complex interaction with gp-130, glycoprotein 130, enables the Janus kinase to activate signal transducer and activator of transcription 3 by phosphorylation (JAK/STAT3 pathway). Accordingly, the STAT3 transcription factor stimulates the expression of inflammatory cytokines. ARDS is promoted by the initiation advancement of the cytokine storm and corresponding cascade (37-39).
Accordingly, the use of IL-6R antagonists may be of great importance as it can hamper the cytokine storm mediated by the IL-6/IL6R complex. The monoclonal antibodies targeting each component of the IL-6/IL-6R complex, including Siltuximab (IL-6 inhibitor), Sarilumab, and Tocilizumab (IL-6R inhibitors), have been suggested for the treatment and mitigation of ARDS symptoms (10, 11). Other agents, including JAK/STAT inhibitors, can interrupt STAT phosphorylation by targeting JAK. Baricitinib is one of the JAK inhibitors currently being investigated for its efficacy and efficiency against COVID-19 (38). Moreover, ruxolitinib, memolitinib, oclacitinib, fedratinib, gandotinib, and pacritinib are other JAK inhibitors, some of which are under clinical trials and may be effective to be employed as agents in combination therapy (40).
3.2.3. Basigin/CD147: A Contributing Receptor for SARS-CoV-2 Cell Entry or Misidentified as a Therapeutic Target?
Basigin (CD147) is a glycoprotein of the immunoglobulin superfamily and is accountable for cancer cell development and enhances viral infections in host tissue (41).
Chen et al. (2005) indicated the importance of Basigin as an enhancer of SARS-CoV invasion (42) and tested and confirmed the interaction of 2SP and Basigin using surface Plasmon resonance and Co-IP assays. Considering the acquired results, they proposed a new potential route for SARS-CoV-2 cell entry, employing Basigin as the associated receptor (41). Several drugs, including Meplazumab, have been suggested to block Basigin, and some of them are being tested in clinical trials (43, 44).
On the other hand, there is controversy if Basigin has significant roles in SARS-CoV-2 cell entry. According to Shilts et al., utilizing Basigin inhibitors as therapeutic options against COVID-19 is not impeccable. They analyzed the binding interaction of 2SP and CD147. Peculiarly, they found no direct interaction between them, pointing to the need for a thorough analysis of its nature before performing clinical trials (45).
3.2.4. CD209L/L-SIGN
L-SIGN (aka CD209L) and DC-SIGN (AKA CD209) receptors belong to the C-type lectin superfamily and are considered as activators of immune response to infectious agents, including mycobacterium tuberculosis (46). On the contrary, they mediate the advancement of the infection and evasion of these pathogens from the immune system, presenting their nature as a double-edged sword. The positive consequence potential of these two receptors is thought to rely on the type of tissue they are expressed in (47). The former is mainly present in dendritic cells and macrophages and the latter in pulmonary endothelial cells and other distinct tissue types. Since both SARS-CoV and SARS-CoV-2 primarily infect alveolar cells, L-SIGN exerts more influence (46).
Previously, the impact of receptors other than ACE-2 on viral cell entry was suggested (especially L-SIGN in SARS-CoV infection) (48). On that basis, research was carried out to investigate the interaction of 2SP with L-SIGN. It was shown that L-SIGN interacts with 2SP-RBD, implying the potential of C-type lectin receptors in the facilitation of SARS-CoV-2 cell entry. Furthermore, the application of soluble CD209L was reported to disrupt this process (49).
4. Conclusions
Considering the absence of a reliable drug, it is crucial to detect COVID-19 infection in suspected cases to prevent further transmissions. Antiviral drugs may be somewhat efficacious against SARS-COV-2, but their solo use for treatment is not efficient enough. Thereby, blocking the agents in the host-SARS-CoV-2 interactome, including TMPRSS2 and signal transducers may greatly reduce its infectivity and fatality. In light of various therapeutic options for treating viral infections, including repurposed drugs, the potential for achieving a tangible result for treatment is still considerable.