1. Background
One of the most common infectious diseases is urinary tract infection (UTI), accounting for more than 150 million cases globally per year (1). It is also considered one of the frequent reasons for referring to medical clinics and is the most common infection after upper respiratory tract infections and gastrointestinal infections (2, 3). Symptoms of recurrent UTI infections include renal hypertension and renal failure if untreated, which can lead to irreversible kidney damage (4). Acute UTI is associated with substantial morbidity and mortality and problems of recurrent infections in both outpatients and hospitalized patients (5).
Bacteria are the main etiologic agent for UTI cases; however, other microorganisms, such as fungi and viruses, can also be rare causes of UTI (6). Escherichia coli is the most prevalent uropathogen and is solely responsible for approximately 70 - 95% of UTIs, followed by other Enterobacteriaceae, such as Klebsiella and Proteus species (7).
The UTIs account for a significant portion of antibiotic consumption in hospitals and out of them, particularly in developing countries where there is no accurate antibiotic prescription control. The UTIs have a large socioeconomic impact and result in antibiotic-resistant strains in the hospital and the community (8). Urinary pathogens have been known to include numerous strains resistant to many of the commonly used antibiotics (9). Multidrug-resistant organisms (MDROs) causing UTIs are widely known as a major threat to the public health system (10).
The MDROs have been increasingly reported in community-acquired and hospital-acquired infections; however, the prevalence is different by region (11). Some bacteria have been recognized as very important in developing MDROs, including extended-spectrum ß-lactamase (ESBL) producing E. coli, Klebsiella spp., methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant (VRE) Enterococcus spp., and multidrug-resistant (MDR) non-fermenting bacteria, such as Acinetobacter spp. and Pseudomonas spp. (12-15). The MDR microorganisms would increase morbidity and mortality rates if they were not covered by suitable antibiotics (16). The UTI manifestation is a widespread reason for referring or returning patients to hospitals in Iran, and unfortunately, the numbers have been rising, according to several studies (17-19).
2. Objectives
The present study aimed to distinguish the bacterial etiological agents from cases of UTI and their drug resistance pattern to commonly used antimicrobial agents among hospitalized patients. It is hoped that the gained information will help provide proper antibiotic therapy for cases of UTI in Tehran, Iran.
3. Methods
3.1. Patents and Sampling
Overall, the current study included 9836 patients referred to a large teaching hospital (Loghman Hospital, Tehran, Iran) from March 2019 to February 2020. These patients were suspected of UTI according to clinical manifestations checked by a specialist. In inpatient or outpatient midstream clean catch, urine samples were collected in a sterile container from each patient. The samples were immediately transferred to a clinical microbiology laboratory, and direct examination and culture tests were performed.
3.2. Bacterial Isolation and Identification Procedures
Media were incubated aerobically at 37ºC for 24 hours, and those cultures which became negative at the end of 24 hours of incubation were further incubated for 48 hours. A sample was considered positive for UTI if a single organism was cultured at 105 cfu/mL (5). The organisms were identified based on phenotypic features, such as gram staining and colony morphology, and biochemical tests. For biochemical analysis, some catalase, oxidase, coagulase, Triple Sugar Iron agar, citrate utilization (Simmons’s citrates medium), urease (Christensen’s Urea Agar), indole, motility, H2S production (Sulfide Indole Motility Medium), methyl red, Voges-Proskauer, Lysine agar, mannitol salt agar, Dnase agar, esculin hydrolysis (Bile-esculin agar), and sugar fermentation tests were considered according to suspected microorganisms.
3.3. Antimicrobial Susceptibility Test
According to the Kirby-Bauer method, antibiotic susceptibility testing was conducted against the most common causative UTI pathogens. The antibiotics used for gram-negative bacteria were trimethoprim/sulfamethoxazole (1.25/23.75 mcg), cefotaxime (30 mcg), cefoperazone (75 mcg), ceftazidime (30 mcg), imipenem (10 mcg), nitrofurantoin (300 mcg), nalidixic acid (30 mcg), amikacin (30 mcg), norfloxacin (10 mcg), ampicillin (10 mcg), and gentamicin (10 mcg), and those used for gram-positive bacteria were trimethoprim/sulfamethoxazole (1.25/23.75 mcg), cefotaxime (30 mcg), cefoperazone (75 mcg), ceftazidime (30 mcg), imipenem (10 mcg), norfloxacin (10 mcg), ampicillin (10 mcg), gentamicin (10 mcg), chloramphenicol (30 mcg), clindamycin (2 mcg), erythromycin (15 mcg), vancomycin (30 mcg), penicillin (10 units), and oxacillin (1 mcg). The results were interpreted according to the recommendation of Clinical and Laboratory Standards Institute criteria as sensitive (S), intermediate (I), and resistant (R) (16).
Reference strains of E. coli, ATCC 25922, Klebsiella pneumoniae, ATCC700603, and P. aeruginosa ATCC 27853 were used as controls for the gram-negative bacteria and included in all daily runs. Staphylococcus aureus ATCC 29213 and Enterococcus faecalis ATCC 29212 were used as gram-positive controls (16). An MDRO isolate was determined as resistant to at least three of the following antimicrobial categories:
Folate pathway antagonists (trimethoprim-sulfamethoxazole), aminoglycosides (amikacin or gentamicin), fluoroquinolones (norfloxacin or nalidixic acid), and nitrofurantoin
3.4. Statistical Analysis
Finally, the chi-square test was used with the help of the SPSS software (version 25.0) for the statistical analyses of this study. A P-value less than 0.05 was considered to be significant.
4. Results
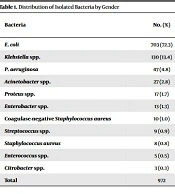
Overall, 972 (9.88%) of 9836 patients with suspected UTI were referred for urine culture. The mean age of the subjects was 35 ± 19.4 years. In addition, 608 (62.6%) female and 364 (37.4%) male subjects were positive in bacterial culture (Table 1). According to the obtained results, females were more susceptible to developing UTIs (P < 0.01). There was no significant correlation between ages with culture results regardless of the microorganisms’ type (P > 0.05) since all mixed infections were excluded from the study. Out of 972 bacterial strains, E. coli was the predominant bacterial isolate and accounted for 703 (72.3%), followed by Klebsiella spp. with 130 (13.4%) and Pseudomonas aeruginosa with 47 (4.8%) isolates (Table 1). Of isolated microorganisms, 96.81% and 3.18% belonged to gram-negative and gram-positive bacteria, respectively. The finding indicated that 83% of the isolates belonged to the six common pathogens, including E. coli, Klebsiella spp., P. aeruginosa, Acinetobacter spp., Proteus spp., and Enterobacter spp.
| Bacteria | No. (%) | Gender | P-Value | |
|---|---|---|---|---|
| Male, No. (%) | Female, No. (%) | |||
| Escherichia coli | 703 (72.3) | 235 (33.4) | 468 (66.6) | 0.0001 |
| Klebsiella spp. | 130 (13.4) | 56 (43.1) | 74 (56.9) | 0.2368 |
| Pseudomonas aeruginosa | 47 (4.8) | 18 (38.3) | 29 (61.7) | 0.2129 |
| Acinetobacter spp. | 27 (2.8) | 19 (70.4) | 8 (29.6) | 0.0992 |
| Proteus spp. | 17 (1.7) | 9 (52.9) | 8 (47.1) | 1.0000 |
| Enterobacter spp. | 13 (1.3) | 9 (69.2) | 4 (30.8) | 0.3179 |
| Coagulase-negative Staphylococcus aureus | 10 (1.0) | 7 (70) | 3 (30.9) | 0.6828 |
| Streptococcus spp. | 9 (0.9) | 3 (33.3) | 6 (66.7) | 0.6828 |
| S. aureus | 8 (0.8) | 3 (37.5) | 5 (62.6) | 0.6792 |
| Enterococcus spp. | 5 (0.5) | 3 (60) | 2 (40) | 1.0000 |
| Citrobacter spp. | 3 (0.3) | 1 (33.3) | 2 (66.7) | 1.0000 |
| Total | 972 | 364 (37.4) | 608 (62.6) | 0.0001 |
Distribution of Isolated Bacteria by Gender
Trimethoprim/sulfamethoxazole showed less activity (n = 550, 56.9%) against commonly isolated pathogens regardless of the type of microorganisms. However, amikacin (n = 787, 84.9%), nitrofurantoin (n = 723, 77.1%), gentamicin (n = 629, 65.6%), imipenem (n = 492, 63.2%), and norfloxacin (n = 577, 61.7%) were observed to be more effective for common pathogens (Table 2). For E. coli as a predominant isolate, 88.4% and 87.5% of the isolates were susceptible to amikacin and nitrofurantoin, respectively. Additionally, 55.5% and 54.1% of the isolates were resistant to nalidixic acid and trimethoprim-sulfamethoxazole, respectively.
| Bacteria | SXT | CTX | CP | CAZ | IMP | FM | NA | AN | NOR | AM | GM |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Escherichia coli | 321 (45.9) | 402 (57.9) | 430 (62.6) | 408 (58.9) | 373 (65.9) | 608 (87.5) | 295 (44.5) | 609 (88.4) | 424 (61.1) | 39 (48.1) | 473 (68.3) |
| Klebsiella spp. | 67 (51.5) | 75 (57.7) | 83 (64.8) | 74 (56.9) | 71 (68.9) | 93 (71.5) | 63 (51.2) | 113 (87.6) | 87 (66.9) | 10 (62.5) | 81 (62.3) |
| Pseudomonas aeruginosa | 10 (21.3) | 26 (55.3) | 40 (85.1) | 37 (78.7) | 31 (79.5) | 7 (14.9) | 9 (19.6) | 36 (78.3) | 39 (83) | 4 (50) | 38 (80.9) |
| Acinetobacter spp. | 5 (18.5) | 4 (14.8) | 5 (18.5) | 4 (14.8) | 3 (14.3) | 3 (11.1) | 4 (15.4) | 6 (22.2) | 6 (22.2) | 1 (25) | 11 (40.7) |
| Proteus spp. | 4 (23.5) | 9 (52.9) | 12 (70.6) | 10 (58.8) | 8 (61.5) | 8 (47.1) | 6 (35.3) | 12 (70.6) | 12 (70.6) | 0(0) | 14 (82.4) |
| Enterobacter spp. | 9 (69.9) | 8 (61.5) | 9 (69.2) | 8 (61.5) | 6 (75) | 4 (30.8) | 8 (61.5) | 11 (84.6) | 9 (69.2) | 1 (33.3 | 12 (92.3) |
| Citrobacter spp. | 1 (33.3) | 2 (66.7) | 2 (66.7) | 2 (66.7) | 2 (66.7) | 3 (100) | 1 (33.3) | 3 (100) | 2 (66.7) | ND | 2 (66.7) |
Frequency/Percentage of Gram-negative Strains Susceptible to Selected Antibiotics a
Antibiotic susceptibility pattern for isolated Klebsiella spp. showed susceptibility to amikacin (87.6%) and nitrofurantoin (71.5%); however, 48.8%, 48.5%, and 43.1% were resistant to nalidixic acid, trimethoprim/sulfamethoxazole, and ceftazidime, respectively. Among P. aeruginosa isolates, as a third common isolated bacteria, cefoperazone (85.1%), norfloxacin (83%), gentamicin (80.9%), and imipenem (79.5%) had the highest rates of susceptibility; however, 85.1%, 80.4%, and 78.7% of the isolates were resistant to nitrofurantoin, nalidixic acid, and trimethoprim/sulfamethoxazole, respectively.
For Acinetobacter spp., only 40.7% of the isolates were susceptible to gentamicin, and a large number of the isolates were resistant to nitrofurantoin (88.9%), ceftazidime (85.2%), and cefotaxime (85.2%). Of 8 isolated S. aureus, 7 (87.5%) were MRSA (oxacillin/methicillin resistant). Only one isolate was susceptible to oxacillin, and one isolate (20%) was detected as VRE Staphylococcus aureus. Among Enterococcus spp., of 5 isolates, 3 (60%) were VRE (Table 3). Of the common isolates collected in this study, 547 isolates (56.27%) were MDR. The Acinetobacter spp. isolates had the highest value (n = 24, 88.8%) for multidrug resistance, followed by 31 (65.9%) of P. aeruginosa, 80 (61.5%) of Klebsiella spp., and 399 (56.7%) of E. coli isolates, respectively (Table 4).
| Bacteria | SXT | CTX | CP | CAZ | IPM | NOR | AM | GM | C | CC | E | V | P | OX |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Staphylococcus aureus | 5 (71.4) | 3 (50) | 5 (83.3) | 2 (100) | 6 (100) | 2 (100) | 1 (100) | 5 (83.3) | 4 (66.7) | 5 (83.3) | 4 (57.1) | 4 (80) | 0 (0) | 7 (12.5) |
| CONS | 7 (70) | 6 (75) | 6 (75) | 2 (66.7) | 7 (100) | ND | 1 (1000) | 7 (87.5) | 7 (87.5) | 8 (88.9) | 5 (50) | 10 (100) | 4 (57.1) | ND |
| Streptococcus spp. | 2 (25) | 5 (71.4) | 4 (57.1) | ND | 3 (42.9) | 1 (100) | 3 (100) | 3 (33.3) | 1 (33.3) | 4 (57.1) | 5 (62.5) | 8 (100) | 4 (80) | ND |
| Enterococcus spp. | 0 (0) | 1 (20) | 1 (25) | ND | 1 (20) | ND | 1 (50) | 1 (25) | 2 (40) | 0 (0) | 1 (20) | 2 (40) | 1 (25) | ND |
Frequency/Percentage of Gram-positive Strains Susceptible to Selected Antibiotics a
| Bacteria | Multidrug Resistance, No. (%) |
|---|---|
| Escherichia coli | 399 (56.7) |
| Klebsiella spp. | 80 (61.5) |
| Pseudomonas spp. | 31 (65.9) |
| Acinetobacter spp. | 24 (88.8) |
| Total | 534 (56) |
Frequency of Bacterial Isolates Showing Multidrug-resistant Phenotype
5. Discussion
Several geographical regions, including Iran, show decreased susceptibility rates to common urinary pathogens; therefore, the global trend to empirically treat community-acquired UTIs might not apply to these regions (20, 21). Antimicrobial resistance in uropathogens should be monitored to improve treatment recommendations. This study was conducted to determine the frequency and antimicrobial susceptibility patterns of community-acquired uropathogens in Iran. The current study demonstrated that E. coli is the most common cause of UTI in Tehran, Iran. This finding corresponds with the data obtained by other investigators (22-24). Klebsiella spp. was the second most common organism, followed by Pseudomonas spp. and Acinetobacter spp., which is similar to a report from Mohammadi-Mehr, and Feizabadi (25) and different from a report from Pouladfar et al. indicating Klebsiella spp. and Enterococcus spp. as prevalent strains next to E. coli (26).
The results of the present study showed that females are more likely to get UTI (P < 0.05) which is similar to nearly all the other reports (17, 19, 24). Fluoroquinolones or nitrofurantoin have been suggested for the empirical treatment of uncomplicated UTIs (27). However, the emergence of high levels of resistance of uropathogenic E. coli against trimethoprim/sulfamethoxazole has been reported in both developing countries (54 - 82%) and developed countries (14.6 - 37.1%) (28-30). The present study also discovered an elevated resistance rate to trimethoprim/sulfamethoxazole (54.1%), which is in accordance with other Iranian studies (19, 24, 31). According to these results, trimethoprim-sulfamethoxazole should no longer be used as the primary empirical treatment in Iran.
The results of the fluoroquinolone susceptibility test in the current study (norfloxacin) showed good action against E. coli (61%), which is in line with other Iranian studies carried out in 2006, indicating constant sensitivity of E. coli isolates to fluoroquinolones. Nitrofurantoin, as the second preferred antibiotic for the treatment of UTI, is effective for the prophylaxis and the treatment of MDR uropathogens in adults, children, and pregnant women. Additionally, it is a relatively safe drug with minimal effects on the resident bowel and vaginal flora (27, 32). Although nitrofurantoin demonstrated better activity against E. coli isolates (87.5% susceptible), it should not be used for serious upper UTIs or for those with systemic involvement (14).
In the present study, Klebsiella spp., as the second common cause of UTI, was resistant to commonly-used antibiotics, except amikacin (87.6%). Therefore, amikacin still remains the best choice for the empirical treatment of severe UTI caused by Klebsiella spp. The susceptibility to norfloxacin has remained constant during the past 3 years. The present study’s sensitivity results (67%) are similar to a previous report from Shenagari et al., with 55% sensitivity (33). This finding might contribute to the limited usage of norfloxacin in Iranian patients.
Considering the current study’s results, P. aeruginosa, with a 5% incidence, was the third most common cause of hospital-acquired UTIs. The currently studied Pseudomonas strains were susceptible to the second-line drugs, such as cefoperazone, norfloxacin, and gentamycin, with more than 80% of cases; however, most of these isolates were associated with high resistance to the first-line used antibiotics, namely nitrofurantoin, trimethoprim/sulfamethoxazole, and nalidixic acid. These findings are in agreement with another Iranian report in which 80% of isolates were sensitive to norfloxacin, and only 11% were sensitive to trimethoprim/sulfamethoxazole (34). Increased susceptibility was observed for nitrofurantoin, nalidixic acid, and trimethoprim/sulfamethoxazole, compared to the results of a previous study (35). This status might contribute to the reduced use of these antibiotics in Iran.
Acinetobacter spp. is known to be important in nosocomial UTIs (36). Acinetobacter spp. isolates demonstrated high resistance to most antibiotics, such as nitrofurantoin, imipenem, ceftazidime, cefotaxime, norfloxacin, ampicillin, trimethoprim/sulfamethoxazole, and nalidixic acid with an average of 83%. Despite most reports and in agreement with the results of studies by Rahimi and Rezaie Keikhaie et al., Acinetobacter spp. isolates showed partly good sensitivity to gentamicin (40%) (37, 38). In contrast to the results of the current study, Mortazavi et al. (39) reported very high resistance to gentamicin and amikacin simultaneously among 80 A. baumannii strains from Ahvaz, south-west Iran.
Previous studies reported the prevalence of S. aureus among UTI patients from 0.8%, 1%, and 6.92 to 11.65% (24, 40-42). The present study’s results showed that 0.8% of patients were infected with S. aureus. The current study showed a high resistant rate (87%) to methicillin/oxacillin (MRSA) in comparison to previous national reports in 2012 (48%) and 2015 (28%) (43, 44). This difference might be related to the non-suitable usage of antibiotics or the low number (5) of studied organisms. A significant increase in MDR pathogenic strains to different antibiotics has been reported worldwide (45). Accordingly, 534 MDR (56%) isolates were detected. Of the 534 MDR isolates, 399 (57%) were E. coli. A lower percentage of MDR E. coli (63%) was found in Poland (22%) and Venezuela (25%) among isolates from community-acquired and hospital-acquired UTIs (7, 46). This diversity in MDR frequency reflects differences in antibiotic prescription and infection control policies in any region worldwide. In conclusion, a relatively high frequency of bacterial resistance was observed in the urine samples collected from Loghman Hospital in Tehran.
The data also indicated that most isolated microorganisms belonged to gram-negative bacilli (97%), and E. coli was the most frequent agent of UTIs (72.3%) in the current study. Considering bacterial diversity causing UTIs, aminoglycosides, such as amikacin, are recommended as the first choice, and nitrofurantoin as the second choice for the treatment of UTIs in Tehran. Nalidixic acid and trimethoprim/sulfamethoxazole, due to reduced efficiency against UTI causative agents, are no longer suggested for the empirical therapy of UTI.
Despite the precious findings on the resistance rate of uropathogens, there are some limitations in the present work. The major drawback pertained to the retrospective design of the study and the inability to have access to the patient’s health records; therefore, the authors were unable to analyze and report the patients’ demographic data and the correlation between the risk factors and underlying pathologies conditions with UTIs. In addition, the lack of molecular characterization of the resistance determinants in the studied isolates and no detection of ESBLs are other limitations of the current study. Further studies are essential to monitoring the rate of bacterial resistance among UTI patients in other hospitals in Iran.
5.1. Conclusions
The potential antimicrobial resistance is one crucial consideration for physicians when selecting an antibiotic for the treatment of infectious diseases, particularly for patients with UTIs. In most cases, antimicrobial chemotherapy is often empiric and should be determined by identifying the most common etiological agents and their antimicrobial susceptibility profiles.