1. Background
Antibiotic resistance is a growing problem in clinical centers throughout the world. This condition may limit treatment options and increase patient mortality and morbidity (1, 2). Incorrect and unnecessary usage of antibiotics and, in some circumstances, prophylaxis with broad-spectrum antibiotics increase the risk of antibiotic resistance in different societies (3). Various reports in Iran show different antibiotic-resistant patterns for causative microorganisms (4-7). Gram-negative bacteria show resistance to the third and fourth generations of cephalosporins, which can be important for the treatment of this type of bacteria (8). Carbapenem antibiotics are used as selected antibiotics for the treatment of these resistant bacteria. Unfortunately, there are reports on carbapenem resistance in Pseudomonas aeruginosa, Acinetobacter baumannii, and Klebsiella pneumoniae as nosocomial isolates from all over the world (9). Some A. baumannii and P. aeruginosa organisms isolated from wound and burn infections are resistant to all antibiotics except colistin, and they can be designated as human "red alarm" pathogens (10, 11).
Methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant S. aureus (VRSA) are the most critical gram-positive nosocomial organisms, and their treatment is a big challenge for clinicians (12). Different reports of the ESKAPE bacteria, including Enterococcus spp., Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp. have been published (4-9, 12). The Centers for Disease Control and Prevention (CDC) announced the ESKAPE bacteria as life-threatening microorganisms worldwide (13). All these antibiotic-resistant bacteria are in the World Health Organization’s (WHO) priority group for which new antibiotics are urgently needed (14).
Some studies report the presence of the ESKAPE bacteria in Iran, which could subsequently cause complicated therapeutic problems and increase mortality and morbidity, as in other countries (6, 7, 10).
2. Objectives
This study's objective was to identify antibiotic-resistant patterns of bacteria containing multidrug-resistant (MDR) P. aeruginosa and A. baumannii, third-generation resistant E. coli, K. pneumoniae, Enterobacter spp. MRSA, VRSA, and vancomycin-resistant Enterococcus spp. (VRE) based on the Clinical and Laboratory Standards Institute (CLSI) 2015 and the WHO global antimicrobial resistance surveillance system (Glass).
3. Methods
3.1. Setting and Bacterial Isolates
In this cross-sectional study, the positive culture of E. coli, K. pneumoniae, Enterobacter spp., P. aeruginosa, A. baumannii, S. aureus, and Enterococcus spp. was gathered from nine provinces, including Mashhad, Hamedan, Tabriz, Tehran, Sanandaj, Zahedan, Golestan, Esfahan, and Ahvaz, in Iran between May 2016 and March 2017.
Selected bacteria from the culture in each city were collected from different clinical samples like blood, urine, CSF, bronchial samples, and wound. The collected bacteria were sent to the Pediatric Infections Research Center (PIRC) laboratory. All bacteria were identified according to conventional biochemical and microbiological methods such as colony morphology observation, gram stain, oxidase, triple sugar iron (TSI), DNase, and mannitol.
3.2. Categorized Bacteria into Five Groups of ESKAPE Bacteria
The five groups of the ESKAPE bacteria that are life-threatening include E. coli, K. pneumoniae, and Enterobacter spp. resistant to 3rd generation (3rdG) cephalosporin, according to the CLSI definition; MDR P. aeruginosa and A. baumannii: Based on the CDC guideline; MRSA: S. aureus resistant to cefoxitin 30µg; VRSA: Vancomycin-resistant S. aureus; and VRE: Vancomycin-resistant Enterococcus spp. (15, 16).
3.3. Antibiotic Susceptibility Testing
Antibiotic susceptibility testing was carried out using Kirby-Bauer disc diffusion testing according to the Clinical and Laboratory Standards Institute guidelines (15) against ampicillin/sulbactam (10/10 µg), piperacillin-tazobactam (100/10 µg), cefepime (30 µg), imipenem (10 µg), meropenem (10 µg), gentamicin (10 µg), amikacin (30 µg), tobramycin (10 µg), ciprofloxacin (5 µg), trimethoprim/sulfamethoxazole (1.25/23.75 µg), and minocycline (30 µg) for gram-negative bacteria, and E. coli ATCC 25922 was used as a control strain in the antibiotic susceptibility testing. Erythromycin (15 µg), clindamycin (2 µg), ciprofloxacin (5 µg), doxycycline (30 µg), and trimethoprim/sulfamethoxazole (1.25/23.75 µg) were prepared for the S. aureus antibiotic test. Moreover, the use of S. aureus ATCC 2921375, considered the control in the minimum inhibitory concentration (MIC) method in the susceptibility of vancomycin, was evaluated by determining the MIC in S. aureus. Ciprofloxacin (5 µg), ampicillin (10 µg), gentamicin (120 µg), and linezolid (30 µg) were used to determine the antibiotic susceptibility in Enterococcus spp.
The antibiotic disks used in this study were purchased from MAST Company (Mast Diagnostics, UK), England, and Muller Hinton agar was purchased from BD Company, USA.
3.4. Statistical Analysis
Statistical data processing was performed using statistical package for the social sciences (SPSS) v. 22.0 (IBM SPSS Statistics) to determine the percentage of antibiotic resistance or susceptibility.
4. Results
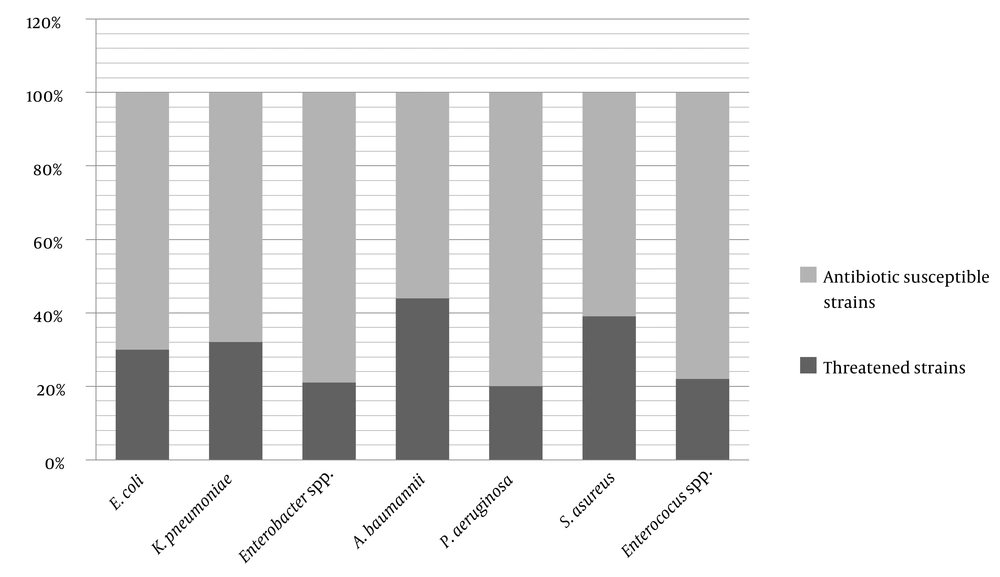
This study collected 5522 bacteria from clinical specimens in microbiology laboratories in nine provinces in Iran. Based on the inclusion criteria, 1666 bacteria, including E. coli, K. pneumoniae, Enterobacter spp. resistant to 3rd generation cephalosporin, MDR P. aeruginosa and A. baumannii, MRSA, and VRE, were identified and involved in this study. The specimens were gathered from 568 (46%) females and 658 (53%) males. The median age was 43 ± 29 y. The frequency of multiple antibiotic-resistant bacteria is shown in Table 1.
| E. coli | K. pneumonia | Enterobacter spp. | P. aeruginosa | A. baumannii | S. aureus | Entrococcus spp. | Total | |
|---|---|---|---|---|---|---|---|---|
| Number of Bacteria | 1222 | 696 | 621 | 675 | 879 | 615 | 814 | 5522 |
| Threatened Bacteria | 370 (30) | 222 (32) | 130 (21) | 136 (20) | 388 (44) | 241 (39) | 179 (22) | 1666 (30) |
a Values are expressed as No. (%) unless otherwise indicated.
The result of the different antibiotic susceptibility of the selected antibiotic-resistant isolates is shown in Table 2 and Figure 1.
| SAM | PTZ | FEP | IMI | MER | GM | AK | TO | CIP | SXT | MIN | |||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | S | I | R | |
| 3rd G. Cephalosporin Resistant E. coli | 14 | 9 | 77 | 94 | 4 | 2 | 84 | 5 | 11 | 57 | 6 | 38 | 91 | 2 | 7 | 35 | 3 | 62 | 22 | 1 | 77 | ||||||||||||
| 3rd G. Cephalosporin Resistant K. pneumoniae | 8 | 4 | 89 | 50 | 11 | 39 | 49 | 8 | 43 | 37 | 1 | 62 | 56 | 2 | 42 | 37 | 4 | 60 | 18 | 1 | 81 | ||||||||||||
| 3rdG. Cephalosporin-Resistant Enterobacter spp. | 4 | 4 | 92 | 37 | 20 | 43 | 32 | 26 | 43 | 17 | 1 | 82 | 38 | 9 | 54 | 23 | 48 | 29 | 16 | 3 | 81 | ||||||||||||
| MDR P. aeroginosa | 20 | 10 | 70 | 2 | 1 | 97 | 10 | 3 | 87 | 7 | 1 | 92 | 3 | 4 | 93 | 15 | 4 | 81 | 4 | 3 | 94 | 6 | 0 | 94 | 3 | 3 | 94 | ||||||
| MDR A. baumannii | 6 | 10 | 84 | 1 | 0 | 99 | 0 | 1 | 99 | 0 | 1 | 99 | 0 | 1 | 99 | 8 | 3 | 88 | 7 | 1 | 93 | 13 | 2 | 85 | 3 | 1 | 96 | 4 | 3 | 92 | 38 | 9 | 53 |
Abbreviations: SAM, ampicillin-sulbactam; PTZ, piperacilline-tazobactam; FEP, cefepime; IMI, imipenem; MER, meropenem; GM, gentamicin; AK, amikacin; TO, tobramycin; CIP, ciprofloxacin; SXT, co-trimoxazole; MIN: minocycline.
Piperacillin-tazobactam and minocycline are the most effective antibiotics for MDR P. aeruginosa and A. baumannii, with 20% and 38% susceptibility rates, respectively (Table 2). Amikacin is the most effective antibiotic for K. pneumoniae and Enterobacter spp., with 56% and 38% susceptibility rates, respectively. For E. coli, imipenem was the most effective antibiotic, with 94% susceptibility rate (Table 2). Resistance patterns of MRSA and Enterococcus spp. are shown in Tables 3 and 4, respectively.
| E | CD | CIP | DOX | SXT | |
|---|---|---|---|---|---|
| Resistance | 75 | 59 | 68 | 37 | 30 |
| Intermediate | 14 | 11 | 8 | 24 | 4 |
| Sensitive | 11 | 30 | 24 | 39 | 66 |
Abbreviations: E, erythromycin; CD, clindamycin; CIP, ciprofloxacin; DOX, doxyciclin; SXT, trimethoprim/sulfamethoxazole.
| CIP | AM | GM (120) | LZD | |
|---|---|---|---|---|
| Resistance | 98 | 81 | 100 | 16 |
| Intermediate | 0.4 | _ | _ | 6 |
| Sensitive | 1.6 | 19 | _ | 78 |
Abbreviations: CIP, ciprofloxacin; AM, ampicillin; GM, gentamycin; LZD, linezolid.
5. Discussion
The ESKAPE bacteria, announced by CDC, are a considerable global health problem because infection by them can increase morbidity and mortality (13). These bacteria can prolong the hospitalization time and increase the imposition of costs on the health system and patients (6, 7, 10). A. baumannii is one of the most prevalent ESKAPE causes of infection and tends to become MDR isolates by obtaining antibiotic resistance plasmids. Unfortunately, 44% of A. baumannii strains were MDR based on our definition in this study. A systematic review in South East Asia in 2018 showed that 58.51% of A. boumannii isolates from the intensive care unit (ICU) were MDR (17). The higher rate of MDR A. baumannii in a review in South East Asia compared to our results can be due to the different source of isolated bacteria. In our study, the specimens were collected from different clinical isolates, but in the review article, only ICU isolates were surveyed.
In a study in Kermanshah, Iran, in 2017, 50% of A. bommanii isolated from ICU were MDR and extensive drug-resistant (XDR) (18). In a study in 2017 on BAL specimens from Babul, 47.9% of the specimens were A. baumannii, and 91.4% were MDR. Owlia et al. showed high antibiotic resistance on MDR-resistant A. baumannii in Iran (19). These different rates in detecting MDR strains may be because of different kinds of specimens, populations, definitions, andquality of used antibiotic discs.
A study by Musyoki et al. on A. baumannii showed that 85% MDR A. baumannii and amikacin were the most effective antibiotics (20). However, our results showed minocycline as the most effective antibiotic. The difference between the two studies can be because of using different antibiotic therapy protocols in the two countries.
A study in Morocco in 2019 on MDR A. baumannii showed that colistin and co-trimoxazole were the most effective antibiotics. However, it is notable that the susceptibility to minocycline was not evaluated (21).
Our results showed that the presence of 20% of P. aeruginosa was MDR. However, in a Chinese study in 2018, 15% of isolated P. aeroginosa were MDR (22). The higher rate of identified MDR P. aeruginosa in our study may be because of using different antibiotic stewardship in these two countries and the different sources of specimens. Vahdani et al. and Lari et al. confirmed higher rates of resistance to all tested antibiotics except colistin (6, 23). It is foreseeable that we will have more antibiotic resistant bacteria in samples collected from burn patients because of the increased use of broad-spectrum antibiotics. In another study in Jordan on respiratory specimens, 21.5% of organisms were P. aeruginosa, and 52.5% were MDR (24).
Zarei-Yazdeli et al. revealed that 75% of isolates were MDR and that ciprofloxacin was the most effective antibiotic (25). A study in Egypt in 2019 confirmed 66.6% MDR P. aeruginosa strains and showed that imipenem had a low resistance rate (26). Also, in India in 2017, imipenem was the most active antibiotic for MDR P. aeruginosa (27). Different rates of MDR P. aeruginosa and different effective antibiotics in MDR strains indicate the need for antibiotic stewardship in healthcare to consider the most effective antibiotics for treating MDR strains of P. aeruginosa.
Our results showed 30% resistance to 3rdG cephalosporin in isolated E. coli. In a study in Saudi Arabia, in 2018, of clinical isolates, 49.5% was resistant to cefotaxime in E. coli specimens (28). In a study in Nepal in 2017 from blood cultures, 84.5% of isolated E. coli were resistant to 3rdG cephalosporins (29). A report from the heart ICU in Tehran in 2017 showed that all E. coli bacteria were resistant to cefotaxime (30). These differences in the results may be because of the source specimens of our study, as most E. coli were isolated from urine and different antibiotic usage in these countries.
The results of antibiotic susceptibility testing indicated that 32% of K. pneumoniae was resistant to the 3rd generation cephalosporin. Moremi et al. showed that 38.5% of K. pneumonia was resistant to 3rdG cephalosporin isolated from burn wounds in India (31). Various antibiotic therapies and sources of specimens can lead to different results in different studies. Mamishi et al. reported 29% cefotaxime-resistant K. pneumoniae isolated from different clinical specimens (32). This result is almost similar to ours, and it can be a similar antibiotic usage in Iran to treat K. pneumoniae infection. In our study, 3rdG, cephalosporin-resistant K. pneumoniae showed resistance to cefepime (89%) and, consequently, to co-trimoxazole (81%). In Yazdansetad et al.'s study, co-trimoxazole in 3rd generation cephalosporin-resistant strains had the highest resistance (33), similar to the results of our study. Ferreira et al., in their study on K. pneumoniae isolated from ICU patients showed that 3rdG cephalosporin-resistant strains were ESBL (34).
Our study showed that 21% of Enterobacter spp. was resistant to 3rdG cephalosporin. In a study in Germany in 2019, 21% of collected Enterobacter spp. was resistant to 3rdG cephalosporin (35). In a study on E. cloacae in nosocomial infections in Tehran (2012), 23% of bacteria were resistant to these groups of antibiotics (36), similar to our results. The reason is the proximity of antibiotic therapy patterns in these two studies. A study in Ethiopia in 2018 showed that 79.6% of gram-negative bacteria were resistant to ceftazidime (37), which is a higher rate compared to our results. The reason is the extensive use of this antibiotic in Ethiopia.
The resistance pattern of S. aureus in the north, west, and south of Iran showed that 53.7%, 40.27%, and 43% were MRSA, respectively (38-40). This result is higher than our result by 27%, which might be due to consuming materials and testing the antibiotic susceptibility process. Our results showed that 68% and 59% were resistant to ciprofloxacin and clindamycin, respectively. Kaur and Chate showed 100% resistance to ciprofloxacin and 97% to clindamycin in collected MRSA (41). The results of the Indian study are higher than ours probably because of the different rate of antibiotic consumption in the two countries. Co-trimoxazole is the most effective antibiotic on MRSA strains in our study.
The rate of VRE collected in our study was 22%. In a study in 2018 in the northwest of Iran, the prevalence of VRE was 18.75% in different clinical specimens (42). In a study in 2017 in Saudi Arabia, the VRE rate was 17% (43). These frequencies are lower than ours, resulting from different methods and consumption materials used in these two studies. Also, a different pattern of antibiotic usage in the two countries could be another reason. In a study from two centers in Tehran on VRE cases in children with acute lymphoblastic leukemia, the VRE prevalence was 25%, similar to our results (44). Similarly, Armin et al. showed low resistance to linezolid (7). In our study, a high percentage of VRE cases was susceptible to linezolid (78%).
5.1. Conclusions
This study's results can be used as antibiotic stewardship in the considered hospitals in this research. The rate of antibiotic resistance in most bacteria causing nosocomial infection can be a guide in the experimental administration of antibiotics. This data can help physicians use more correct antibiotics to treat infectious patients. Moreover, it can be an alarmingly high rate of emerging bacteria in a selected hospital in Iran, and it may be a warning to stop misusing of broad-spectrum antibiotics.
On the other hand, the emergence of these resistant strains shows high resistance to available antimicrobial agents. Thus, we are encountering a limited choice of antibiotics, which may become narrower in the future. These results give valuable information in strategic planning for antibiotic prescription, especially in empiric therapy.