1. Background
Septicemia in neonates is an important bacterial blood infection with clinical symptoms (1). It is one of the most dangerous and common neonatal diseases worldwide. The lack of complete development of the immune system is one of the causes of this disease in neonates (2, 3). Sepsis is more common among low-birth weight and preterm neonates than term infants (4). The incidence rate of sepsis among very low-birth-weight neonates in neonatal intensive care units (NICUs) may reach 30% (5).
Despite recent advances in medical science and the use of appropriate guidelines for treating infections in preterm neonates, septicemia remains one of the leading causes of death in preterm neonates in the world (6). Population-based studies in developing countries have shown that the incidence rate of this clinical infection has a range of 49 - 170 cases per 1000 live births (7, 8). Neonatal mortality is one of the key driving factors for assessing community health (9-11). The prevalence of blood infections in neonates is about 1 - 4 cases per 1000 infants. Different measures, such as proper care during pregnancy, observance of hygiene principles by neonatal ward staff, proper hand washing before contact with any neonate, and proper use of antibiotics, could control the prevalence of sepsis in preterm neonates (12).
Neonatal septicemia is a common infectious disease that happens during the first four weeks of a newborn's life and is characterized by clinical and laboratory findings along with a positive blood culture (13-16). The causes of sepsis vary across geographical regions and over time (17). Even within one country, the identified causes of sepsis in medical centers may differ. In developed countries, the most common organism causing sepsis was Group A Streptococcus (Streptococcus hemolyticus) in the 1930s and 1940s. This bacterium was overtaken by Staphylococcus aureus in the 1980s. Since the late 1980s, Escherichia coli and Klebsiella have been responsible for approximately 60%-70% of neonatal sepsis cases. However, in these countries, the causes of sepsis have varied over time across different geographical regions (11).
Various studies have investigated the causes of neonatal sepsis in developing countries. In these studies, Gram-negative bacteria have been reported as the primary cause of sepsis. For example, Pseudomonas and Klebsiella have been the primary causes of neonatal sepsis in India (18). Different organisms are involved in neonatal sepsis in various geographical areas. In a study by Stoll et al., Group B Streptococcus was found to be the most common cause of sepsis in the US; however, in non-US countries, S. aureus was identified as the most common causative agent of sepsis (19).
In a one-year study investigating sepsis causes in NICUs in Australia, the most common causes of late-onset sepsis were Group B Streptococcus and coagulase-negative Staphylococcus (CONS) (20). In another study by Motara and colleagues, out of 103 hospitalized neonates, 91 cases were diagnosed with late-onset sepsis, among which CONS were reported in 63 cases (21). Moreover, in another research conducted on 406 hospitalized neonates, 41.6% of blood culture-positive infants were found to be infected by S. aureus (15.4%) and K. pneumoniae (65.4%) (7). The number of preterm births in Iran is increasing, and hospitalization of preterm infants immediately after birth is inevitable in most cases (22, 23).
Febrile infants who appear abnormal, with decreased peripheral perfusion, hyperventilation, restlessness, lethargy, and abnormal heart rate, are strongly suspected of having septicemia. Physical examinations and history taking may determine the origin of the disease (5, 24). The initial symptoms are similar to those of other infectious diseases. Some signs and symptoms include chills, hypotension, tachycardia, hyperventilation, and decreased peripheral vascular resistance. Purpuric and petechial lesions in the form of skin rashes are typical for meningococcemia (i.e., meningococcal septicemia caused by Neisseria meningitidis). Certain skin ulcers, called bull’s eye lesions, are caused by Pseudomonas aeruginosa. Disseminated intravascular coagulation could be manifested with purpura and bleeding at the site of blood sampling. Moreover, hypotension can lead to lactate acidosis, anuric renal failure, and peripheral gangrene (25).
Antibiotics are the most common medications administered in NICUs; however, their excessive and long-term use is accompanied by an increased incidence of necrotizing enterocolitis and mortality rate in neonates (26). The treatment method for neonatal sepsis is based on standard guidelines, supportive care, and antibiotic prescription (27). Antibiotics can alter the gastrointestinal microbial flora of infants and possibly make them susceptible to infections caused by opportunistic organisms, leading to the emergence of antibiotic-resistant pathogens (28). This resistance to antibiotics may, in turn, lead to the failure of neonatal sepsis treatment and ultimately cause irreparable damage to the infant (29). Bacterial agents are the leading causes of neonatal septicemia, and the use of antibiotics leads to the emergence of antibiotic-resistant bacteria. In addition, the incidence rate of septicemia varies in different communities (30).
Considering the disability of preterm infants admitted to NICUs and their urgent need for further monitoring, this study aimed to identify the bacterial agents causing neonatal septicemia and determine their antibiotic sensitivity patterns according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. Bacterial agents have been reported as the main leading causes of neonatal septicemia at different times and places. Infants are at risk of infections caused by resistant organisms due to long-term hospitalization in the NICU and exposure to various invasive procedures and interventions (31). Therefore, continuous epidemiological surveillance in hospitals for infection control, especially in the NICU, requires antibiotic guidelines based on the relevant microbial agents. The NICU of Al-Zahra Hospital in Tabriz is the largest NICU in East Azerbaijan province. Therefore, most mothers with high-risk childbirths in this region are sent to this center.
2. Objectives
This study was conducted to determine the prevalence rate and antibiotic susceptibility patterns of bacterial agents causing neonatal septicemia.
3. Methods
This descriptive cross-sectional research was conducted on 1000 infants hospitalized in NICUs 1 and 2 and the neonatal ward of Al-Zahra Medical Center in Tabriz from March 2019 to June 2020. The sample size required to achieve the study objectives was 1000 neonates (95% confidence level and 0.9 test power).
3.1. Inclusion Criteria
Inclusion criteria for mothers and neonates in Al-Zahra Medical Center based on a self-report method included not using alcohol or drugs, having a neonate with a gestational age of 26 - 39 weeks, and a neonatal weight of 800-3800 g, not having congenital malformations, and breastfeeding.
3.2. Exclusion Criteria
Exclusion criteria entailed formula feeding, non-cooperation of the mother, and delivery at any center other than Al-Zahra Medical Center.
3.3. Sample Collection
Data were collected using a researcher-made questionnaire composed of two parts. The first part contained questions designed to collect information about the infant's demographic characteristics, such as age, weight, gender, and gestational age (the mother's gestational age was determined based on the date of her last menstrual period and the results of obstetric ultrasound), birth weight, delivery type (cesarean delivery or vaginal delivery), and first- and fifth-minute Apgar scores. Apgar is a rapid assay conducted on a neonate one and 5 minutes after birth. These 2 scores show how well the neonate has endured the delivery process and how well the neonate is doing outside his/her mother's womb. The second part of the questionnaire included questions about the causes of infection, antibiotics administered to the infant, bacterial strains and their drug resistance patterns, and neonatal blood culture results according to laboratory reports, which were completed in the questionnaire after follow-up by the researcher (30). A self-report researcher-made questionnaire was prepared based on the previous valid studies available in the scientific literature and the checklist of the Ministry of Health and Medical Education in Iran. The content validity of this questionnaire was evaluated using the content analysis method with the participation of 10 faculty members and specialists in the neonatal specialty at Tabriz University of Medical Sciences. Necessary modifications were made based on their viewpoints after obtaining the approval of the Ethics Committee and the Research Council of Tabriz Pediatric Research Center. Before implementing the plan, informed consent was received from all mothers with hospitalized infants. The study was executed at Tabriz Pediatric Research Center after obtaining the approval of the Ethics Committee with the code IR.TBZMED.REC.1398.576.
Bacterial culture is one of the methods commonly used to detect infectious agents. This test should be performed before antibiotic administration for all patients with suspected sepsis. Therefore, venous blood samples (1 mL) were taken from all neonates at hospitalization and inoculated in a brain heart infusion medium if they had clinical signs and symptoms of sepsis. In the next step, the blood sample was incubated at 37°C for 24 hours in an incubator and checked daily for small colonies of growing bacteria. Next, all culture media were incubated at 37°C for one week. Afterward, all negative cultures were maintained for another week. Gram-positive bacteria were identified using catalase, coagulase, novobiocin sensitivity, growth on mannitol salt agar medium, and DNase tests.
Gram-negative bacteria were also recognized by culturing on selective media and using differential biochemical tests, namely triple sugar iron agar, sulfide-indole-motility, urea agar, methyl red-Voges Prauskauer (MRVP), and Simmons citrate agar. Bacillus spp., Micrococcus spp., and Corynebacterium spp. were considered contaminants. If the neonates met the following three criteria, they were diagnosed with septicemia: (1) Treatment with specific antibiotics for at least seven days; (2) having at least one positive blood culture; and (3) having clinical symptoms of sepsis, including increased need for a ventilator, augmented frequency of bradycardia and/or apnea, temperature instability, sudden collapse, rapid deepening of jaundice, and feeding difficulties (vomiting, abdominal distension, and gastric retention).
3.4. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed for all positive culture samples using the dispensing method based on CLSI principles (2019) by performing the Kirby-Bauer disk diffusion susceptibility assay. For this purpose, 3 - 5 pure bacterial colonies were collected using a sterile wire needle and placed into a normal saline tube or nutrient broth to make turbidity equivalent to 0.5 McFarland turbidity standard. Next, the microbial suspension was cultured on Mueller-Hinton agar medium using a sterile swab. The antibiotic disks used in this method included ceftriaxone (30 µg), cloxacillin (1 µg), amikacin (10 µg), and ampicillin (10 µg) for Gram-positive bacteria and cefotaxime (30 µg), ceftazidime (30 µg), ciprofloxacin (5 µg), gentamicin (10 µg), imipenem (10 µg), cotrimoxazole (1.25/23.75 µg), amikacin (10 µg), and piperacillin/tazobactam (100/10 µg) for Gram-negative bacteria (Padtanteb, Iran). Antibiotic susceptibility patterns were evaluated by comparing the inhibition zones with the CLSI 2019 guidelines.
3.5. Statistical Analysis
In order to determine the scientific reliability of the questionnaire, a pilot study was conducted, and two researchers and 20 subjects completed the questionnaire. Afterward, according to Cohen’s Kappa agreement coefficient of 0.89, the study was conducted using this questionnaire. The obtained data were entered into SPSS software version 22 and analyzed using the Chi-square test. Laboratory constant instruments were used for all infants. Before conducting the research, all instruments were calibrated, and a calibration certificate was installed on each instrument.
4. Results
Based on the data of 1000 infants hospitalized in the neonatal ward and NICUs, this study showed that 78 cases (7.8%) had positive blood cultures. The average age of the participants in this research was 33.5 weeks (standard deviation: 3.0), with a range of 25 - 36 weeks of gestation. The mean weight of the study participants was 2137 g (standard deviation: 324). Demographic data, including the infant's gender and age, maternal age, delivery type, birth weight, and blood group, are presented in Table 1.
| Variables | Total, No. (%) | Sepsis Cases, No. (%) | P-Value |
|---|---|---|---|
| Gender of infant | 0.12 | ||
| Boy | 568 (56.8) | 46 (58.9) | |
| Girl | 432 (43.2) | 32 (41) | |
| Age of infants, d | 0.03 | ||
| Under 5 | 203 (20.3) | 21 (26.9) | |
| 5 - 10 | 312 (31.2) | 24 (30.7) | |
| 10 - 20 | 248 (24.8) | 19 (24.3) | |
| Over 20 | 237 (23.7) | 14 (17.9) | |
| Age of mother, y | 0.04 | ||
| Under 20 | 278 (27.8) | 19 (24.3) | |
| 20 - 30 | 425 (42.5) | 34 (43.5) | |
| Over 30 | 297 (29.7) | 25 (32) | |
| Type of delivery | 0.12 | ||
| Natural | 316 (31.5) | 27 (34.5) | |
| Cesarean | 686 (68.5) | 51 (65.5) | |
| Birth weight, kg | 0.12 | ||
| Less than 1 | 163 (16.3) | 12 (15.4) | |
| 1 - 2 | 387 (38.7) | 31 (39.7) | |
| More than 2 | 450 (45.0) | 35 (44.8) | |
| Infant blood type | 0.23 | ||
| A | 282 (28.2) | 24 (30.76) | |
| B | 248 (24.8) | 18 (23) | |
| AB | 91 (9.1) | 7 (8.97) | |
| O | 379 (31.9) | 29 (37.2) | |
| First-minute Apgar | 0.13 | ||
| Less than 6 | 450 (45) | 31 (39.74) | |
| More than 6 | 550 (55) | 47 (60.25) |
According to the results shown in Table 2, the most common microorganisms causing neonatal blood infections were CONS (46.1%), K. pneumoniae (28.2%), S. aureus (7.7%), E. coli and S. agalactiae (5.1%), and Acinetobacter baumannii and P. aeruginosa (3.8%). The highest bacterial resistance was observed to ampicillin and gentamicin (nearly 100%). Furthermore, the highest susceptibility of Gram-positive strains was to ceftriaxone (Table 3), and the highest susceptibility of Gram-negative strains was to piperacillin/tazobactam, imipenem, and ciprofloxacin (Table 4).
| Bacteria | No. (%) |
|---|---|
| Staphylococcus coagulase neg. | 36 (46.15) |
| Staphylococcusaureus | 6 (7.7) |
| Streptococcus agalactia | 4 (5.12) |
| Klebsiellapneumoniae | 22 (28.2) |
| Escherichia coli | 4 (5.12) |
| Pseudomonas aeruginosa | 3 (3.85) |
| Acinetobacterbaumannii | 3 (3.85) |
| Antibiotic | Staphylococcus Coagulase Negative, n = 36 (46.15%) | Staphylococcus aureus, n = 6 (7.7%) | Streptococcus agalactia, n = 4 (5.12%) | |||
|---|---|---|---|---|---|---|
| Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | |
| Ceftriaxone (30 µg) | 19 (52.7) | 17 (47.3) | 2 (33.3) | 4 (66.7) | 0 (0) | 4 (100) |
| Cloxacillin (1 µg) | 26 (72.2) | 10 (27.8) | 4 (66.6) | 2 (33.4) | 0 (0) | 4 (100) |
| Amikacin (10 µg) | 13 (36.1) | 23 (63.9) | 3 (50) | 3 (50) | 4 (100) | 0 (0) |
| Ampicillin (30 µg) | 36 (100) | 0 (0) | 6 (100) | 0 (0) | 1 (25) | 3 (75) |
a Values are expressed as No. (%).
| Antibiotic | Klebsiella pneumonia, n = 22 (28.2%) | Escherichia coli, n = 4 (5.12%) | Pseudomonas aeruginosa, n = 3 (3.8%) | Acinetobacter baumannii, n = 3 (3.8%) | ||||
|---|---|---|---|---|---|---|---|---|
| Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | |
| Ampicillin (10 µg) | 22 (100) | 0 (0) | 4 (100) | 0 (0) | 3 (100) | 0 (0) | 3 (100) | 0 (0) |
| Amikacin (10 µg) | 20 (90.9) | 2 (9.1) | 2 (50) | 2 (50) | 2(66.6) | 1 (33.4) | 3 (100) | 0 (0) |
| Cefotaxime (30 µg) | 22 (100) | 0 (0) | 1 (25) | 3 (75) | 2 (66.6) | 1 (33.4) | 3 (100) | 0 (0) |
| Ceftazidime (30 µg) | 22 (100) | 0 (0) | 2 (50) | 2 (50) | 3 (100) | 0 (0) | 3 (100) | 0 (0) |
| Piperacillin/ Tazobactam (100/10 µg) | 0 (0) | 22 (100) | 0 (0) | 4 (100) | 0 (0) | 3 (100) | 0 (0) | 3 (100) |
| Ciprofloxacin (5 µg) | 18 (81.8) | 4 (18.2) | 1 (25) | 3 (75) | 1 (33.4) | 2 (66.6) | 1 (33.4) | 2 (66.6) |
| Cotrimoxazole (1.25/23.75 µg) | 22 (100) | 0 (0) | 2 (50) | 2 (50) | 2 (66.6) | 1 (33.4) | 3 (100) | 0 (0) |
| Gentamicin (10 µg) | 22 (100) | 0 (0) | 3 (25) | 1 (75) | 3 (100) | 0 (0) | 3 (100) | 0 (0) |
| Imipenem (10 µg) | 5 (22.8) | 17 (77.2) | 0 (0) | 4 (100) | 0 (0) | 3 (100) | 1 (33.4) | 2 (66.6) |
a Values are expressed as No. (%).
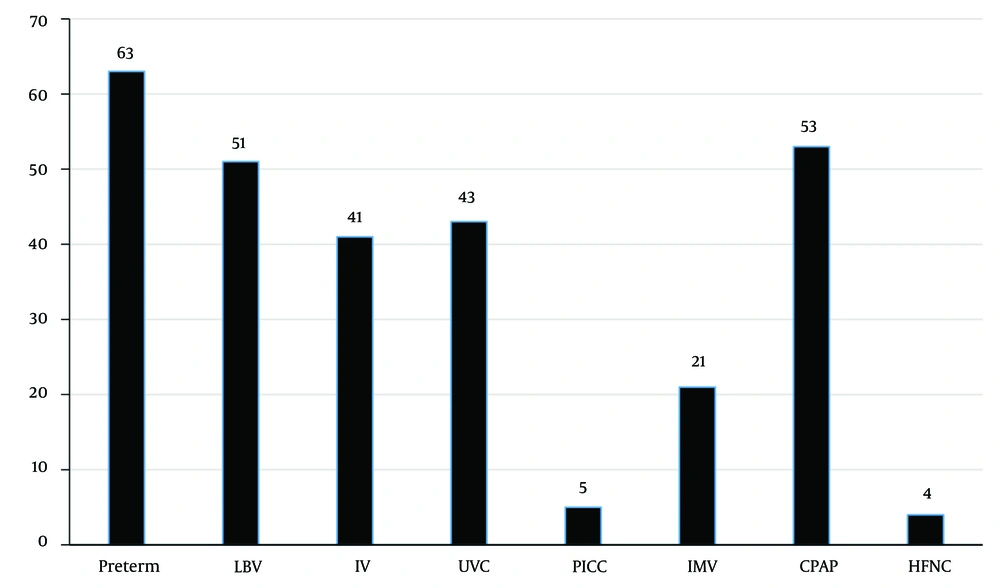
In the present study, prematurity (80.7%), continuous positive airway pressure (68%), and low birth weight (65.4%) were the most common contributing factors to neonatal infection (Figure 1).
5. Discussion
Our results indicated that the bacterial culture of the blood samples of 7.8% of the neonates hospitalized in Al-Zahra Educational and Remedial Center was positive. The results of 78 cases were consistent in determining the role of common bacterial agents in neonatal blood infection. All hospitalized newborns were included in this study. Given that most neonates were term and less likely to be hospitalized, the infection rate is reduced in infants if they are admitted and manipulated aggressively. The results of the present study are consistent with the findings of Rashidi et al., reporting a prevalence rate of 17.9% for sepsis among 700 neonates (32). The lower prevalence of septicemia in the present study than in the latter investigation could be attributed to the difference in the geographical areas where bacterial strains causing neonatal sepsis were isolated. The prevalence of sepsis was reported to be 46.6% in another study by Eyesus et al., which is also higher than the current research findings (11, 32, 33).
The most common microorganisms causing neonatal blood infection were CONS (46.1%) (Gram-positive) and Klebsiella (28.2%) (Gram-negative). In a study by Eyesus et al., the percentage of CONS and Klebsiella was 40.8% and 15.8%, respectively, which is in line with the findings of the present study (33). Rashidi et al. reported that CONS (75%) was found to be the main leading cause of neonatal infection (32). In other similar studies conducted in recent years in Canada (30), India (28), Mexico (16), Australia (14), Nigeria (34), and Taiwan (25), CONS and Klebsiella have been reported as the most common Gram-positive and Gram-negative bacterial causes of neonatal blood infection, respectively. In research by Hosseini et al., Klebsiella and Acinetobacter were reported as the most common bacterial causes of neonatal infection. This result about Klebsiella agrees with the current study (27). In addition, Eyesus et al. found that Klebsiella and S. aureus were the most common bacterial causes of sepsis in infants (15.8% and 40.8%, respectively) (33). Moreover, our results are in line with the findings of another recent study conducted by Hosseini et al. at Al-Zahra Educational and Remedial Center in Tabriz (7, 27, 35-38).
The highest bacterial resistance levels observed in this study were related to ampicillin and gentamicin (nearly 100%). On the other hand, the highest susceptibility of Gram-positive strains was to ceftriaxone, and the highest susceptibility of Gram-negative strains was to piperacillin/tazobactam, imipenem, and ciprofloxacin. These results are in line with the findings of another study by Mollahosseini et al., in which Gram-negative strains were sensitive to ciprofloxacin. In their study, sensitivity to gentamicin was reported among the isolates (1.3%) (36), while in the present study, the highest resistance after ampicillin was to gentamicin. According to the hospital guidelines, the first antibiotics used in the NICU for all neonates without symptoms of sepsis are ampicillin and gentamicin, which leads to the emergence of antibiotic resistance. In line with the results of the current study, Eyesus et al. observed antibiotic resistance to ampicillin (37%). In addition, Hosseini et al. reported that the highest resistance was to ampicillin (47.9%) (27, 33, 36). Although antibiotic resistance patterns are always similar in one country, they may change in different studies due to different gene transfer mechanisms. Another reason may be increasing antibiotic therapy in some areas that can change susceptibility in bacteria.
In this research, male infants were 1.5 times more likely to have sepsis than female infants, which is consistent with the results of Eyesus et al., showing that the prevalence of sepsis was 59.1% in males and 41.9% in females (33). Moreover, Heydarian et al. reported that the prevalence of sepsis in males was twice higher than in females (39). This difference could be attributed to gender-related genes stimulating the immune system (33, 36).
Since 2012, Klebsiella has become resistant to common antibiotics prescribed to treat neonatal sepsis. Considering the common bacterial causes of neonatal infections, recent studies have indicated the urgent need for annual surveys to track and detect resistant strains. It has been estimated that 30 - 40% of neonatal deaths worldwide (a total of 4,035,000 deaths in 2001, according to the World Health Organization) are associated with infectious diseases. Even in developed countries, there are significant challenges for obstetricians and neonatologists in managing infectious diseases during pregnancy and in newborns (27).
5.1. Conclusions
Considering the increased prevalence of CONS infections shown in the present and previous studies, it is possible to reduce the mortality and disability caused by these infections by proper antibiotic therapy based on accurate antibiogram results, providing proper prenatal care to prevent the birth of preterm infants, as well as observing health principles in delivery, neonatal ward, and NICU. Therefore, due to different microbial factors and drug resistance patterns in diverse regions, annual surveys should be conducted to determine drug resistance patterns, emphasizing preventive measures so that a minimum number of neonates develop sepsis. The results of the current study may assist physicians in selecting appropriate antibiotics for treating neonatal sepsis.