1. Background
The fecal microbiota of each person is unique (like a fingerprint), which is formed during birth and influenced by the mother's microbiota, living environment, nutrition, and lifestyle. The first 2 to 3 years of life are the most important time for the development of a healthy microbiota. Natural birth and feeding the baby with breast milk create and maintain a healthy gut microbiome; however, exposure to toxins and drugs, especially antibiotics and environmental pollution, underlying diseases, and hospitalization can affect the composition of the intestinal microbiota a lot (1). Widespread and inappropriate use of antibiotics has led to a significant increase in antibiotic-resistant bacteria around the world. One of the most important resistances among gram-negative bacteria is related to the occurrence of strains encoding beta-lactam inactivating enzymes, especially broad-spectrum beta-lactamases. The gastrointestinal tract is a place for the horizontal spread of resistance genes among gram-negative pathogens and members of the natural intestinal microbiota (2).
Enterobacteriaceae is a common member of the intestinal microbiota, which could cause extraintestinal infections in patients during hospital stays. Among the resistance mechanisms in Enterobacteriaceae, extended-spectrum beta-lactamase (ESBL) has shown a high prevalence in patients of different age groups (1, 2). Children are very vulnerable to infections caused by multidrug-resistant (MDR) Enterobacteriaceae, mainly due to the limited possibility of prescribing broad-spectrum antibiotics for prophylaxis and therapeutic regimens. Therefore, it is necessary to know about the distribution and establishment of ESBL-producing Enterobacteriaceae (ESBL-E) in children of each community. Previous studies have shown the increasing trend of this resistance phenotype in pediatric infections, which had a direct relationship with the mortality of patients (3, 4). Members of ESBL-E have been reported in various regions worldwide. Despite their role in hospital-acquired infections, their isolation from community-acquired infections has now become a major challenge for infection control programs (3-5). Escherichia coli and Klebsiella pneumoniae from the Enterobacteriaceae family are currently considered the most common MDR pathogenic bacteria worldwide, showing their clinical importance (5, 6).
Studies examining ESBL-E colonization in neonates and pediatrics have shown a frequency of 4.3% to 75% (7-9) and 18.5% to 57.5%, respectively (10-12). However, transmission in the community is lower, ranging between 0.1% and 12.4% (13, 14). Several studies have shown an increase in the prevalence of ESBL-E colonization in the intestine of children (East Africa, 42%; Pakistan, 40%; Israel, > 50%; China, 46%; America, 4%; and Germany, 12%) (15, 16). Further research is needed to understand the factors contributing to the changes in the distribution and occurrence of ESBL-E in children. These factors can include the socioeconomic status of a community and the strategy for the management of antibiotic prescription in hospitals, the community, and the food chain. In the hospital setting, risk factors such as the presence of intravascular catheters, surgery, prolonged stay in the intensive care unit (ICU), crowded hospital rooms, and transmission via contaminated hands of hospital staff are known to significantly increase the risk of intestinal colonization with ESBL-E in children.
2. Objectives
Considering the importance of screening tests for ESBL-E in the intestinal tract and its impact on preventing endogenous hospital infections, the current study aimed to investigate the primary frequency of ESBL-E in stool samples of children upon admission to the pediatric ICU (PICU) and its alteration at discharge time. Due to the limited information available on the hospital and environmental reservoirs for the transmission of ESBL-E to children, as well as the potential role of antibiotic administration in promoting their growth, this study conducted phenotypic and molecular screening tests on the most prevalent ESBL-E members from each patient and investigated their homology between different patients.
3. Methods
3.1. Patients and Sampling
This cross-sectional study included 80 of the 90 patients admitted to the PICU at Mofid Children's Hospital in Tehran between July 2021 and December 2021. Rectal swab samples were collected from each patient upon admission to the PICU and at discharge time, which was 48 hours after of PICU stay. The study was approved by the Ethics Committee of the Research Institute of Children's Health, Shahid Beheshti University of Medical Sciences, Tehran, Iran (code: IR.SBMU.RICH.REC.1399.055). All experimental stages of the study were performed in the Microbiology Laboratory of Pediatric Infections Research Center in the Mofid Children's Hospital Research Center. The sampling was performed according to the ethical principles for medical research involving human subjects. Accordingly, permission from the children's parents was obtained using an approved consent form. Patient information, including age, sex, type of illness, history of hospitalization, duration of hospitalization, and medication, were collected using a questionnaire. Patients who were hospitalized for less than 48 hours or those who were referred from other hospitals with a history of > 48-hour hospitalization were excluded from the study.
3.2. Characterization of Enterobacteriaceae in Stool Samples
To isolate and identify Escherichia species, Klebsiella spp., and Enterobacter spp., rectal swab samples were cultured on MacConkey agar medium (Beckton Dickinson and Company, Sparks, MD, USA). Diagnostic tests (such as the analysis of colony morphology, Gram staining, oxidase test, and biochemical reactions in triple sugar iron agar (TSI), Simon citrate agar, sulfide indole motility medium (SIM), methyl red-Voges-Proskauer broth (MR/VP), and urease media) were investigated. The bacterial strains E. coli ATCC 25922 and Klebsiella pneumoniae ATCC 1706 were used as standards for all tests. Fresh colonies of each confirmed isolate were stocked in a tryptic soy broth medium (Merck, Germany) containing glycerol (15%, volume/volume) until their use for antimicrobial susceptibility testing. Members of Enterobacteriaceae that differ from the noted genera and non-Enterobacteriaceae isolates were excluded from the study.
3.3. Antimicrobial Susceptibility Test and ESBL Phenotype
Antimicrobial susceptibility testing was performed on pairs of the same bacterial species isolated from each patient upon admission and discharge. Antibiotic susceptibility to imipenem (10 µg), trimethoprim-sulfamethoxazole (1.25 - 23.75 µg), cefotaxime (30 µg), cefazolin (30 µg), amoxicillin-clavulanic acid (10 - 20 µg), cefepime (30 µg), ciprofloxacin (5 µg), meropenem (10 µg), ceftriaxone (30 µg), gentamicin (10 µg), and ampicillin (10 µg) (Beckton Dickinson and Company, Sparks, MD, USA) was performed by the disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) standard M100 protocol (17). The double-disc synergy test (cefotaxime (30 µg) and cefotaxime-clavulanic acid (10 - 30 µg)) was done for all the isolates that were resistant to ceftazidime. A ≥ 5-mm increase in a zone diameter of cefotaxime in combination with clavulanic acid compared to the zone diameter of the agent when tested alone was considered a possible ESBL phenotype. Due to the limitation for screening of the ESBL-E isolates using ceftazidime and ceftazidime-clavulanic acid discs, further confirmation was done by polymerase chain reaction (PCR) using specific primers, as described below. Multidrug-resistant, extensively drug-resistant (XDR), and pan-drug-resistant (PDR) phenotypes were determined for each ESBL-E to isolate according to the study of Magiorakos et al. (18).
The detection of ESBL-associated genes was performed using specific primers for blaCTX-M, blaSHV, blaPER, blaVEB, and blaTEM. The nucleotide sequences of the primers and PCR conditions are shown in Table 1. The products were analyzed after electrophoresis on an agarose gel. Positive control strains carrying related β-lactamase genes were kindly provided by Dr Fereshteh Fani.
| Gene | Primers (5′-3′) | Product Length (bp) | Source |
|---|---|---|---|
| blaCTX-M | 909 | (19) | |
| F | GAGTTTCCCCATTCCGTTTC | ||
| R | CAGAATAAGGAATCCCATGGTT | ||
| blaTEM | 860 | (20) | |
| F | TCAACATTTCCGTGTCG | ||
| R | CTGACAGTTACCAATGCTTA | ||
| blaSHV | 930 | (21) | |
| F | TTTATCGGCCYTCACTCAAGG | ||
| R | GCTGCGGGCCGGATAACG | ||
| blaPER | 925 | (22) | |
| F | ATGAATGTCATTATAAAAG | ||
| R | AATTTGGGCTTAGGGCAGAA | ||
| blaVEB | 645 | (23) | |
| F | TTGGACTCTGCAACAAATACGC | ||
| R | CGACTTCCATTTCCCGATGC | ||
| ERIC-PCR | Variable | (24) | |
| F | ATGTAAGCTCCTGGGGATTCAC | ||
| R | AAGTAAGTGACTGGGGTGAGCG |
Nucleotide Sequences of the Primers Used in This Study
3.4. Phylogenetic Analysis and Molecular Typing
The identified isolates with the same antibiotic resistance pattern in the first and second samples of the patient were typed using enterobacterial repetitive intergenic consensus-PCR (ERIC-PCR) using specific primers (Table 1). A combined phenetic and phylogenetic dendrogram was drawn using NTSyS software version 2.02 to show the relatedness of the isolates for each genus using numerical data of the ERIC-PCR banding pattern, carriage of ESBL encoding genes, and resistance phenotype to antibiotics.
3.5. Statistical Analysis
Chi-square and Fisher's statistical tests were used to evaluate qualitative variables. All analyses were performed using SPSS version 22 (SPSS Inc, Chicago, IL, USA). P values less than 0.05 were considered statistically significant.
4. Results
The average age of the studied patients was 45.71 ± 40.22 months, ranging from 1 to 204 months (48 (60%) boys and 32 (40%) girls). Of the 80 patients, 9 had a history of previous hospitalization, and the average duration of previous hospitalization was 2.27 ± 2.5 days, with a range of 1 to 7 days. The reasons for PICU admission were mainly related to brain/neurological disorders, neurosurgery, general surgery, infections, cancer, and lung, digestive, endocrine, heart, and blood diseases. A recent history of antibiotic therapy was recorded for cefotaxime (2%), ceftriaxone (3.8%), vancomycin (3.8%), clindamycin (2%), ciprofloxacin (2%), meropenem (2%), and other antibiotics (7.69%). Biochemical tests confirmed the presence of Enterobacteriaceae in all 160 pairs of samples collected from 80 patients during the first and second phases of their hospitalization in the PICU (100%). Of these, 70 (88%) were associated with Escherichia spp., 16 (20%) were associated with Klebsiella spp., and 14 (18%) were associated with Enterobacter spp. Also, of the 80 investigated cultures in the second phase, 58 (73%) were associated with Escherichia spp., 20 (25%) were associated with Klebsiella spp., and 17 (21%) were associated with Enterobacter spp. Of the 160 cases, 104 (52/80) were selected for further investigation based on having similar bacteriology results in terms of bacterial species between the first and second phases of sampling.
The highest frequency of Enterobacteriaceae antibiotic resistance in the first phase was observed in cephazolin (80.7%), amoxicillin-clavulanic acid (82.6%), and ampicillin (82.6%), and in the second phase, it was observed in cephazolin (86.5%), amoxicillin-clavulanic acid (80.7%), and ampicillin (88.4%). In both the admission and discharge phases, Enterobacter spp. and Klebsiella spp. showed more similar patterns of antibiotic resistance and a higher frequency of resistance to imipenem, meropenem, gentamicin, and cefotaxime-clavulanic acid antibiotics compared to Escherichia spp. isolates. The Klebsiella spp isolates were also resistant to ciprofloxacin at a higher percentage compared with Escherichia spp. and Enterobacter spp. at both periods. No significant difference was detected in the frequency of MDR strains between the primary and second stool samples in PICU (82.6% vs 86.5%, respectively). The presence of XDR strains was observed in Escherichia spp. (83.7% vs. 76.7%, respectively) and Klebsiella spp. (100% vs. 100%, respectively) more than Enterobacter spp. isolates (57.1% vs 57.1%, respectively). More details about antimicrobial susceptibility results are presented in Tables 2 and 3.
| Antibiotic Resistance Among the Fecal Enterobacteriaceae | Admission % (n) | Discharge % (n) | P Value |
|---|---|---|---|
| Imipenem | 19.2 (10/52) | 21.1 (11/52) | 0.999 |
| Escherichia spp. | 13.9 (6/43) | 11.6 (5/43) | 0.999 |
| Enterobacter spp. | 42.8 (3/7) | 71.4 (5/7) | 0.999 |
| Klebsiella spp. | 50 (1/2) | 50 (1/2) | 0.999 |
| Trimethoprim-sulfamethoxazole | 69.2 (36/52) | 65.3 (34/52) | 0.66 |
| Escherichia spp. | 72 (31/43) | 67 (29/43) | 0.62 |
| Enterobacter spp. | 57.1 (4/7) | 57.1 (4/7) | 0.999 |
| Klebsiella spp. | 50 (1/2) | 50 (1/2) | 0.999 |
| Cefotaxime-clavulanic acid | 25 (13/52) | 32.6 (17/52) | 0.51 |
| Escherichia spp. | 18.6 (8/43) | 27.9 (12/43) | 0.44 |
| Enterobacter spp. | 57.1 (4/7) | 57.1 (4/7) | 0.999 |
| Klebsiella spp. | 50 (1/2) | 50 (1/2) | 0.999 |
| Cephazolin | 80.7 (42/52) | 86.5 (45/52) | 0.38 |
| Escherichia spp. | 76.7 (33/43) | 83.7 (36/43) | 0.38 |
| Enterobacter spp. | 100 (7/7) | 100 (7/7) | 0.999 |
| Klebsiella spp. | 100 (2/2) | 100 (2/2) | 0.999 |
| Amoxicillin-clavulanic acid | 82.6 (43/52) | 80.7 (42/52) | 0.999 |
| Escherichia spp. | 79 (34/43) | 76.7 (33/43) | 0.999 |
| Enterobacter spp. | 100 (7/7) | 100 (7/7) | 0.999 |
| Klebsiella spp. | 100 (2/2) | 100 (2/2) | 0.999 |
| Cefotaxime | 71.1 (37/52) | 76.9 (40/52) | 0.49 |
| Escherichia spp. | 67.4 (29 /43) | 74.4 (32/43) | 0.47 |
| Enterobacter spp. | 85.7 (6/7) | 85.7 (6/7) | 0.999 |
| Klebsiella spp. | 100 (2/2) | 100 (2/2) | 0.999 |
| Cefepime | 44.2 (23/52) | 48 (25/52) | 0.28 |
| Escherichia spp. | 44.1 (19/43) | 46.5 (20/43) | 0.47 |
| Enterobacter spp. | 42.8 (3/7) | 57.1 (4/7) | 0.999 |
| Klebsiella spp. | 50 (1/2) | 50 (1/2) | 0.999 |
| Ciprofloxacin | 32.6 (17/52) | 38.4 (20/52) | 0.67 |
| Escherichia spp. | 32.5 (14/43) | 39.5 (17/43) | 0.64 |
| Enterobacter spp. | 28.5 (2/7) | 28.5 (2/7) | 0.999 |
| Klebsiella spp. | 50 (1/2) | 50 (1/2) | 0.999 |
| Meropenem | 15.3 (8/52) | 19.2 (10/52) | 0.79 |
| Escherichia spp. | 11.6 (5/43) | 13.9 (6/43) | 0.999 |
| Enterobacter spp. | 28.5 (2/7) | 42.8 (3/7) | 0.999 |
| Klebsiella spp. | 50 (1/2) | 50 (1/2) | 0.999 |
| Ceftriaxone | 71.1 (37/52) | 73 (38/52) | 0.82 |
| Escherichia spp. | 67.4 (29/43) | 72 (31/43) | 0.63 |
| Enterobacter spp. | 85.7 (6/7) | 71.4 (5/7) | 0.999 |
| Klebsiella spp. | 100 (2/2) | 100 (2/2) | 0.999 |
| Gentamicin | 26.9 (14/52) | 23 (12/52) | 0.82 |
| Escherichia spp. | 20.9 (9/43) | 18.6 (8/43) | 0.999 |
| Enterobacter spp. | 57.1 (4/7) | 42.8 (3/7) | 0.999 |
| Klebsiella spp. | 50 (1/2) | 50 (1/2) | 0.999 |
| Ampicillin | 82.6 (43/52) | 88.4 (46/52) | 0.57 |
| Escherichia spp. | 79 (34/43) | 86 (37/43) | 0.57 |
| Enterobacter spp. | 100 (7/7) | 100 (7/7) | 0.999 |
| Klebsiella spp. | 100 (2/2) | 100 (2/2) | 0.999 |
Comparison of Changes in the Antibiotic Resistance of Isolates of Enterobacteriaceae Family During Admission to and Discharge from the Pediatric Intensive Care Unit a
| Admission % (n) | Discharge % (n) | P Value b | AB Usage c | P Value d | |
|---|---|---|---|---|---|
| MDR | |||||
| Enterobacteriaceae | 82.6 (43/52) | 86.5 (45/52) | 0.78 | ||
| Escherichia spp. | 79 (34/43) | 84 (36/43) | 0.78 | ||
| Enterobacter spp. | 100 (7/7) | 100 (7/7) | 0.999 | ||
| Klebsiella spp. | 100 (2/2) | 100 (2/2) | 0.999 | ||
| XDR | |||||
| Enterobacteriaceae | 80 (42/52) | 75 (39/52) | 0.63 | ||
| Escherichia spp. | 83.7 (36/43) | 76.7 (33/43) | 0.58 | ||
| Enterobacter spp. | 57.1 (4/7) | 57.1 (4/7) | 0.999 | ||
| Klebsiella spp. | 100 (2/2) | 100 (2/2) | 0.999 | ||
| PDR | |||||
| Enterobacteriaceae | 1.9 (1/52) | 9.6 (5/52) | 0.20 | ||
| Escherichia spp. | 2 (1/43) | 7 (3/43) | 0.61 | ||
| Enterobacter spp. | 0 (0/7) | 28 (2/7) | 0.46 | ||
| Klebsiella spp. | 0 (0/2) | 0 (0/2) | 0.999 | ||
| ESBL | |||||
| Enterobacteriaceae | 48 (25/52) | 42.3 (22/52) | 0.69 | ||
| Escherichia spp. | 51 (22/43) | 46 (20/43) | 0.83 | Metronidazole e | 0.016 |
| Enterobacter spp. | 28 (2/7) | 14 (1/7) | 0.999 | ||
| Klebsiella spp. | 50 (1/2) | 50 (1/2) | 0.999 |
The Frequency of Multidrug-Resistant, Extensively Drug-Resistant, Pandrug-Resistant, and Extended-Spectrum β-Lactamase Phenotypes Among the Enterobacteriaceae Isolates Upon Admission to and Discharge from the Pediatric Intensive Care Unit a
The frequency of ESBL-E strains was 48% (25/52) upon admission and 42% (22/52) at the time of discharge, which did not show a significant difference. Among the bacterial genera under investigation, the most common ESBL-E was Escherichia spp. during admission and discharge (31% and 34%, respectively).
No significant correlation was detected between the prescribed antibiotics, except for meropenem (P = 0.04), which showed a significantly higher resistance in children who received this antibiotic, and the frequency of resistance to the antibiotics. Also, the prescription and use of broad-spectrum antibiotics (except metronidazole confirmed in the case of ESBL Escherichia spp. strains) did not show a correlation with the colonization of ESBL-E in hospitalized patients. No significant difference was found in different age groups and the gender of patients between the frequencies of ESBL-E.
Among the ESBL genes investigated upon admission and discharge, the most common gene causing the ESBL phenotype was blaCTX-M (92% vs. 81%, respectively), followed by blaSHV (28% vs 32%, respectively) and blaTEM (28% vs 14%, respectively). In addition, blaTEM and blaSHV genes were identified in Escherichia and Enterobacter genera, and the blaCTX-M gene was present in all 3 studied bacterial genera. Further, blaPER and blaVEB genes were not detected in the studied samples.
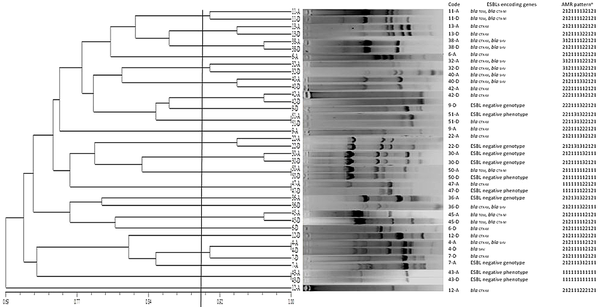
The results of the phylogenetic analysis showed that most of the ESBL-E isolates at the time of discharge had the same origin as the colonized isolates at the time of admission. As was shown in Figure 1, for ESBL Escherichia spp., in terms of drug resistance patterns, ERIC-PCR genotype, and types of ESBL genes, 5 distinct patients showed 100% homology in the clonality of their fecal isolates upon admission and discharge (patient No. 51, 50, 47, 4, and 43). In terms of drug resistance and ERIC-PCR patterns, 9 patients showed more than 90% homology (patient No. 11, 13, 38, 32, 40, 42, 22, 30, and 45), and 2 patients showed less than 90% homology between the isolates in the samples collected upon admission and discharge (patient No. 36 and 7). Most of the isolates (84.2%, 16/19) from each patient showed high phylogenetic homology with their discharge counterparts. The possibility of cross-transmission between patients was shown in only 1 case (patient No. 7) with a phylogenetic homology of higher than 90% with the admission and discharge samples of patient No. 4. This homology was not confirmed at the molecular level due to the different banding pattern.
Homology in the phenetic and molecular patterns of Escherichia species isolates from the stool of pediatric patients in the pediatric intensive care unit upon admission and discharge. The scale bar shows the percentage of homology. Phylogenetic relationship of Escherichia spp. strains isolated from the primary and secondary stool samples of children hospitalized in the pediatric intensive care unit (PICU) based on antimicrobial sensitivity and enterobacterial repetitive intergenic consensus-polymerase chain reaction (ERIC-PCR) patterns. Dendrogram was drawn using numerical data using NTSyS software version 2.02. Strains with 100% similarity were considered identical, while others with more than 90% similarity were defined as related strains. a, antimicrobial resistance (AMR) patterns show the similarity of antibiotic susceptibility phenotypes among isolates. The numerical codes represent patterns of resistance (1), susceptible (2), and intermediate (3) to different antibiotics orderly as follows: Imipenem, trimethoprim-sulfamethoxazole, cefotaxime-clavulanic acid, cefazolin, amoxicillin-clavulanic acid, cefotaxime, cefepime, ciprofloxacin, meropenem, ceftriaxone, gentamicin, and ampicillin.
5. Discussion
The carriage of ESBL-E in the intestinal tract of non-hospitalized and hospitalized children was reported by other studies. A similar frequency of ESBL-E in the stool samples of outpatient children in comparison to our samples on the admission time was detected (France, 44% (16), Zimbabwe, 52% (25), and Chad, 38% (26)). Extended-spectrum β-lactamas-producing Enterobacteriaceae in the stool samples of children who were hospitalized for ≥ 48 hours showed frequencies of 57%, 59%, 58%, 44%, and 48% in Madagascar (11), Ethiopia (15), Seattle (23), Chad (25), and South Africa (27), respectively, which are in relative agreement with our results. In the present study, Escherichia spp. were characterized as the most common ESBL-E upon admission and discharge, which is consistent with the results of similar studies in Chad (25) and Japan (72% and 79.8%, respectively). Contrary to these results, Klebsiella pneumoniae was reported as the most characterized species among ESBL-E in Madagascar (11), Morocco (28), South America (23), and Ethiopia (26). Although these studies were not conducted on pediatrics, the high frequency of ESBL-E was attributed to the prescription, high consumption of antibiotics, poor hygiene, and high density of patients in the hospital, which is more common in developing countries.
In the present study, the high frequency of blaCTX-M, especially among Escherichia spp. isolates, is consistent with the results of molecular epidemiology studies conducted on ESBL-producing isolates among children in America, Europe, and the Middle East. In a study from Morocco, blaCTX-M was the most common (78%), followed by blaSHV (67%) and blaTEM (43%) (28). Also, in South Africa, the most common ESBL gene was blaCTX-M, which was characterized in 95% (76/80) of ESBL-E isolates from stool samples of admitted children in the PICU (27).
In the present study, no similarity was found in terms of phenotype and genotype of drug resistance between different patients at the time of discharge from the PICU, except for 2 cases that showed similarity in the dendrogram but differed in terms of the ERIC-PCR pattern and antibiotic prescription. This result does not confirm the occurrence of cross-infection among patients, which seems to be due to the short period of follow-up after the hospitalization of children in the PICU. Conducting further studies with a longer follow-up can make the role of the hospital clearer in this situation. The lack of hospital transmission of ESBL-E among children in this study could also be explained due to the time of the study, which was associated with the COVID-19 outbreak when standards of infection control prevention were considered strictly. In a similar study in Korea, a lack of cross-transmission of ESBL-E and ESBL-E. coli was similarly reported (29). In another study in Maryland, fecal carriage of ESBL-producing E. coli was reported in 24% (23.97) of patients, of which cross-transmission between patients was confirmed in only 13% (3.23) of patients in the ICU (30). Also, in another study based on the pulsed-field gel electrophoresis (PFGE) method, of the 50 patients carrying ESBL-E, only 1 case of cross-transmission (E. cloacae) was found in the ICU (31).
Considering that in the present study, there was little difference in the prevalence of ESBL-E strains between the time of admission and discharge, and these isolates were detected in the digestive system at the time of arrival of the patients, it seems that there are environmental sources for the bacterial transmission of ESBL-E. According to this hypothesis, studies conducted in Iran have shown food as a carrier of ESBL-E; thus, environmental factors can be responsible for the presence of these bacteria in the digestive system of children. Further investigation is needed to identify these reservoirs in raw food and understand the mechanisms by which they can be transmitted to children. The widespread use of antibiotics in the agriculture and animal husbandry industry has increased antibiotic resistance. A study conducted in Iran in 2021 on turkey meat showed that 28.5% were infected with the ESBL-producing E. coli strain (32). In another study that aimed to identify different genes of ESBLs in E. coli, fresh and frozen meat samples were examined, revealing a contamination rate of 22.5% in the meat samples. This contamination was more evident in fresh meat samples (33). In 2018, Kheradmand and colleagues reported the presence of ESBL-E and contamination of vegetables, such as lettuce (60%) and spinach (83%) (34). It seems that these reservoirs can play a significant role in ESBL-E bacterial transmission. Due to the high prevalence of ESBL-E in the food materials and the environment and their transmission to the children's digestive system, it is recommended to use prophylactic or preventive methods to control infection with these bacteria, mainly in children who refer to hospitals for medical interventions and those who stay in PICU and other hospital wards for long periods. It is also necessary to separate patients carrying ESBL-E and bacteria with multiple resistance from other patients and strictly monitor the prescription and consumption of antibiotics in hospitals to prevent the development of treatment-resistant infections in these patients during hospitalization (35).
5.1. Conclusions
Extended-spectrum β-lactamas-producing Enterobacteriaceae was present in the gastrointestinal tract of children at the time of the initial visit, and hospitalization for 48 hours did not significantly affect the increase of ESBL-E strains. Further studies are required to prove the relationship between hospitalization and increased incidence of ESBL-E. Our study showed little effect of antibiotic prescription on the increased incidence of ESBL-E; however, children receiving meropenem antibiotics showed significantly higher resistance than those not receiving meropenem antibiotics. Also, the administration and consumption of broad-spectrum antibiotics, such as metronidazole, increased the incidence of ESBL Escherichia spp. strains in hospitalized patients. In our patients, Escherichia spp. were found to be the most prevalent among ESBL-E members, and the most common gene causing ESBL-type resistance was blaCTX-M. The incidence of cross-transmission between patients during hospitalization was not confirmed in this study. Further studies are needed to understand the role of ESBL-E bacteria in extraintestinal infections, particularly bacteremia, as well as to investigate the potential persistence of these bacteria as a permanent component of the gastrointestinal microbiota after hospital discharge.