1. Background
In late December 2019, unusual cases of pneumonia with symptoms resembling viral infections were reported in China, quickly spreading to other regions worldwide (1, 2). In early January 2020, the Chinese Center for Disease Control and Prevention (CDC) identified a new coronavirus in a patient’s throat sample, which was later named the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) by the World Health Organization (WHO) (3, 4). The SARS-CoV-2 has caused millions of deaths due to severe respiratory complications, mainly dyspnea, and other organ failures. Respiratory epithelial cells are the primary targets of SARS-CoV-2, and the clinical syndrome of COVID-19 is classified as a lower respiratory tract disease (5, 6). The widespread expression of angiotensin-converting enzyme 2 (ACE-2) (the SARS-CoV-2 receptor) in extrapulmonary tissues may allow the virus to affect these tissues as well. Alongside common respiratory symptoms, patients also experience additional signs such as chest pain, headache, and notably, gastrointestinal (GI) symptoms. Consequently, a variety of extrapulmonary symptoms, including loss of smell and taste, vomiting and diarrhea, and neurological disorders, may result from this phenomenon (7).
The most common and reliable test for diagnosing COVID-19 has been quantitative PCR, or qPCR, which is performed using nasopharyngeal swabs and other upper respiratory tract samples, including throat swabs or, more recently, saliva. In most individuals with symptomatic SARS-CoV-2 infection, viral RNA can be detectable in nasopharyngeal and/or oropharyngeal swabs from the first day of symptom onset, and the viral load peaks during the first week of symptom onset (8). The detection of the virus in specimens derived from non-respiratory organs, as well as the potential for transmission via alternative routes beyond respiratory droplets, remains a subject of ongoing investigation and is not yet fully elucidated in the scientific literature. Various studies conducted on both adults and children have confirmed the presence of viral genomes and even complete viral particles in the blood of infected individuals (6). Other studies have reported the presence of the virus in feces and urine. A study conducted by Peng et al. on various samples from patients with this respiratory infection confirmed the presence of the SARS-CoV-2 virus in blood, urine, rectal swabs, and throat samples using qPCR testing. This suggests that the virus can cause infection through the respiratory, GI, urinary, and blood systems. Therefore, the diagnosis of COVID-19, especially in asymptomatic individuals, can be evaluated in various samples from the patient (9).
Numerous clinical, epidemiological, case studies, and other reports worldwide have demonstrated that a significant number of patients with COVID-19 may develop neurological symptoms and complications affecting both the central and peripheral nervous systems. These neurological manifestations can occur before, during, and even after the onset of common COVID-19 symptoms (10).
2. Objectives
The present retrospective study aimed to investigate and analyze the biological distribution of SARS-CoV-2 among various samples from hospitalized patients diagnosed with COVID-19, based on clinical features and radiological manifestations, and confirmed through genomic diagnosis via reverse transcription-quantitative polymerase chain reaction (RT-qPCR) testing. The results of RT-qPCR tests conducted on non-nasopharyngeal samples — including BAL, feces, ETT, CSF, and sputum — collected from hospitalized patients at Namazi Hospital in Shiraz since the onset of the pandemic and throughout various peaks of COVID-19 will be analyzed and compared. Furthermore, the clinical and paraclinical symptoms across different patient groups will also be assessed.
3. Methods
3.1. Study Design and Patients
This retrospective study analyzed 1,567 samples from 690 female and 877 male patients, with an average age of 38.72 years. The samples were categorized by type: 550 bronchoalveolar lavage (BAL), 464 endotracheal tube (ETT), 45 fecal, 21 cerebrospinal fluid (CSF), and 487 sputum samples. These were collected from patients at Namazi Hospital between April 2020 and September 2022. Patient information, including age, gender, workplace, exposure risks, and symptoms, was recorded through an electronic questionnaire. Additional data on pregnancy, underlying health conditions (such as diabetes, cardiovascular and respiratory diseases, neuromuscular conditions, and kidney or liver disease), and COVID-19 hospitalization were also documented.
3.2. Sample Collection
Sputum samples were collected in sterile containers and sent to the laboratory. Bronchoalveolar lavage, ETT, and CSF samples were obtained by specialist physicians in operating rooms and transported to the laboratory under cold conditions. Fecal samples from COVID-19 patients were collected as requested by physicians, also maintaining the cold chain. All samples were handled with sterile virus transport mediums containing antifungal and antibacterial agents, and they were not subjected to repeated freezing and thawing.
3.3. Laboratory Assays
Nucleic acid extraction from each sample was performed using the Sinnaclon DNA Extraction Kit (DNPTM) 50T-EX6071, Iran. Quantification was conducted using multiplex TaqMan real-time PCR with specified kits (Pishteh Zob Tab, Iran). Each PCR reaction had a final volume of 25 µL. Extracted nucleic acid specimens were evaluated for SARS-CoV-2 with RT-qPCR using a SARS-CoV-2 RdRp/N gene nucleic acid detection kit (Pishtazteb Diagnostics) and the rotor-gene Q real-time PCR system (QIAGEN, Germany) according to the manufacturer’s instructions. The procedure involved adding 15 µL of PCR master mix into a PCR reaction tube with 10 µL of the processed sample. Reactions were incubated at 50°C for 15 minutes and 95°C for 3 minutes, followed by 40 cycles at 94°C for 10 seconds and 55°C for 40 seconds.
3.4. Statistical Analysis
In this study, the study group was classified using descriptive statistics based on the results of SARS-CoV-2 PCR testing. The categorized variables were presented as numbers and percentages for age, with no missing data reported. The chi-square test was employed to identify statistically significant differences among the categorized variables. The SPSS version 26 was utilized for analysis, with a significance threshold set at P < 0.05. There were no restrictions on age, gender, or type of ward for sample collection from patients undergoing COVID-19 testing in this study.
4. Results
In this study, 1,567 samples were collected from patients and categorized into five groups based on the type of sample (Table 1).
| Samples | Total Samples | Median Age | Female/Male | Percentage of Positive COVID-19 Test |
|---|---|---|---|---|
| ETT | 464 | 32.37 ± 31.13 | 148/316 | 11 |
| BAL | 550 | 40.60 ± 29.92 | 309/241 | 20.7 |
| CSF | 45 | 35.67 ± 30.05 | 29/16 | 17.8 |
| Stool | 21 | 8.57 ± 15.95 | 1/21 | 4.76 |
| Sputum | 487 | 47.28 ± 29.82 | 217/270 | 2.9 |
Abbreviations: ETT, endotracheal tube; BAL, bronchoalveolar lavage; CSF, cerebrospinal fluid.
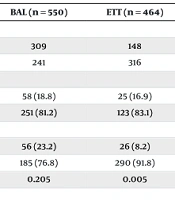
In Table 2, the initial group comprised 464 ETT samples, with 316 obtained from male patients, of whom 26 (8.2%) tested positive, and 148 from female patients, with 25 (16.9%) testing positive. The median age of the group was 16 years. A statistically significant difference in the frequency of the virus was observed in the ETT samples, with females showing a higher positivity rate compared to males (P ≤ 0.05).
| Variables | BAL (n = 550) | ETT (n = 464) | CSF (n = 45) | Stool (n = 21) | Sputum (n = 487) |
|---|---|---|---|---|---|
| Gender | |||||
| Female | 309 | 148 | 29 | 0 | 217 |
| Male | 241 | 316 | 16 | 21 | 270 |
| COVID test results in female; No. (%) | |||||
| Positive | 58 (18.8) | 25 (16.9) | 2 (6.9) | 0 | 4 (1.8) |
| Negative | 251 (81.2) | 123 (83.1) | 27 (93.1) | 0 | 213 (98.2) |
| COVID test results in male; No. (%) | |||||
| Positive | 56 (23.2) | 26 (8.2) | 6 (37.5) | 1 (4.8) | 10 (3.7) |
| Negative | 185 (76.8) | 290 (91.8) | 10 (62.5) | 20 (95.2) | 160 (96.3) |
| P-value | 0.205 | 0.005 | 0.017 | 0.7 | 0.281 |
Abbreviations: BAL, bronchoalveolar lavage; ETT, endotracheal tube; CSF, cerebrospinal fluid.
Among these, 413 samples tested negative for COVID-19 (89%), while 51 samples tested positive (11%). The second group included 550 BAL samples, comprising 241 males and 304 females with a median age of 43.5 years. In this group, 114 patients tested positive for COVID-19 (20.7%). The third group involved patients with various central nervous system (CSF) disorders suspected of having this viral infection, for whom CSF samples were collected for analysis. Out of 45 samples, 29 were from female patients and 16 from male patients, with a median age of 31 years. Eight samples (17.8%) in this group tested positive for COVID-19, while 37 (82.2%) were reported as negative. A statistically significant correlation was observed between gender and the positivity rate in this group, with males showing a higher positivity rate, as 6 males (37.5%) tested positive (P ≤ 0.01).
The fourth group comprised 21 fecal samples taken from patients exhibiting GI symptoms, with a median age of 2 years; all patients in this group were male. Only one fecal sample from these patients (4.8%) tested positive for COVID-19, while the remaining samples (95.2%) were negative. Finally, the presence of the virus in sputum samples was evaluated in a cohort of 487 patients, including 270 males and 217 females. The median age of these patients was 53 years, with 473 samples (97.1%) testing negative for COVID-19 and 14 samples testing positive (2.9%).
Table 3 highlights significant differences in the distribution of negative and positive test results across various sample types and age groups, with all P-values indicating statistical significance. In patients under age 18, BAL and ETT samples show high negative rates (87.7% and 93.8%, respectively), whereas CSF has a notably higher positive rate at 36.8%. Stool and sputum samples show predominantly negative results (95% and 97.7%, respectively). For adult patients, BAL samples have a higher positive rate (25.8%), but sputum remains predominantly negative (96.9%). Notably, CSF in this group has a 96.2% negative rate, highlighting consistent trends across age groups.
| Age Groups and Type of Samples | Total | Negative Test | Positive Test | P-Value |
|---|---|---|---|---|
| 0 - 18 | 0.000 | |||
| BAL | 205 (100) | 180 (87.7) | 25 (12.2) | |
| ETT | 243 (100) | 228 (93.8) | 15 (6.2) | |
| CSF | 19 (100) | 129 (63.2) | 7 (36.8) | |
| Stool | 20 (100) | 19 (95) | 1 (5) | |
| 19 - 103 | 0.000 | |||
| Sputum | 132 (100) | 129 (97.7) | 3 (2.3) | |
| BAL | 345 (100) | 256 (74.2) | 89 (25.8) | |
| ETT | 221 (100) | 185 (83.7) | 36 (16.3) | |
| CSF | 26 (100) | 25 (96.2) | 1 (3.8) | |
| Stool | 1 (100) | 1 (100) | 0 (0) | |
| Sputum | 355 (100) | 344 (96.9) | 11 (3.1) |
Abbreviations: BAL, bronchoalveolar lavage; ETT, endotracheal tube; CSF, cerebrospinal fluid.
a Values are expressed as No. (%).
In this study, 32% (178) of the patients whose BAL samples were analyzed were hospitalized in the intensive care unit (ICU), while similar proportions were observed for patients with ETT samples (32%), CSF samples (32%), fecal samples (47%), and sputum samples (29%) (Table 2). Statistically significant higher levels of positivity were found in BAL samples from ICU patients compared to other sample types (P ≤ 0.0001) (Table 1). Cough (P < 0.05) and dyspnea (P ≤ 0.001) were substantially more common in BAL samples with the viral genome detected than in those that tested negative. In ETT samples, dyspnea was also significantly more common among those with detectable viral genomes. For patients whose fecal samples were evaluated using qPCR, clinical symptoms such as nausea, diarrhea, and body aches were significantly more frequent in those who tested positive for COVID-19 (P ≤ 0.001). It is important to note that in the remaining two groups — those with sputum and CSF samples — no statistically significant differences in clinical symptom occurrence were observed between COVID-19 positive and negative groups.
The majority of patients who had BAL samples taken were those with heart disease (101 patients). Among the patients who had ETT samples taken, the majority were those with chronic neurological diseases, followed by heart disease patients (64 and 58 patients, respectively). For sputum samples, the majority came from heart disease patients (89 patients) (Table 4).
| Variables | BAL | ETT | CSF | Stool | Sputum |
|---|---|---|---|---|---|
| Fever | 141 (25.6) | 107 (23) | 10 (22.2) | 8 (38) | 97 (20) |
| Shortness of breath | 205 (37.3) | 152 (32.8) | 9 (20) | 5 (23.8) | 179 (36.8) |
| Cough | 40 (7.3) | 49 (10.6) | 2 (4.4) | 2 (9.5) | 37 (7.6) |
| Confusion | 82 (85.1) | 48 (10.3) | 2 (4.4) | 1 (4.8) | 65 (13.3) |
| Immune system defects | 9 (1.6) | 10 (2.2) | 0 | 0 | 9 (1.8) |
| Chronic kidney disease | 21 (3.8) | 22 (4.7) | 0 | 2 (9.5) | 13 (2.7) |
| Chronic neurological disease | 76 (13.8) | 64 (13.8) | 9 (20) | 2 (9.5) | 54 (11.1) |
| Chronic lung disease | 13 (2.4) | 4 (0.9) | 0 | 0 | 13 (2.7) |
| ICU hospitalization | 178 (37.6) | 161 (40.9) | 9 (25) | 10 (50) | 146 (35.9) |
| diabetes | 75 (13.6) | 39 (8.4) | 4 (8.4) | 0 | 49 (10.1) |
| Malignancy | 30 (5.5) | 23 (5) | 3 (6.7) | 0 | 34 (7) |
| Heart diseases | 101 (18.4) | 58 (12.5) | 8 (17.8) | 3 (14.3) | 89 (18.3) |
| Liver diseases | 5 (1.1) | 8 (1.5) | 0 | 1 (4.8) | 14 (2.9) |
Abbreviations: BAL, bronchoalveolar lavage; ETT, endotracheal tube; CSF, cerebrospinal fluid; ICU, intensive care unit.
a Values are expressed as No. (%).
5. Discussion
Severe acute respiratory syndrome coronavirus 2 primarily targets the lungs and spreads mainly through the respiratory system. However, since the ACE-2 receptor, which the virus uses to enter cells, is also present in other organs, the virus may infect other body parts. Thus, non-respiratory transmission routes should be considered (11, 12). Using multiple sample types can help prevent viral transmission through non-respiratory routes, such as oral-fecal or via bodily fluids (13). Understanding RNA shedding across various biological samples is crucial for ensuring biosafety and protecting healthcare workers. Additionally, to improve COVID-19 diagnosis and minimize viral spread, it is important to examine the link between disease severity and the presence of the virus in different patient samples (14).
This study focuses on examining the presence of the SARS-CoV-2 genome in fecal, BAL, ETT, and CSF samples from COVID-19 patients and explores how it relates to the severity of their clinical symptoms. Understanding the viral genome’s detection in various clinical samples is crucial for assessing its correlation with disease severity. Our findings revealed that 12% of the 1,567 clinical samples tested positive for SARS-CoV-2, with BAL samples demonstrating the highest positivity rate at 20.7%. This indicates that BAL is a particularly effective sample type for detecting the virus in COVID-19 patients.
Today, molecular diagnostic tests to identify the causes of pneumonia (bacterial, viral, and fungal) from BAL samples are widely available. BAL is an excellent method for diagnosing lung infections, particularly in immunocompromised patients. Studies have shown that the detection rate of various microorganisms in BAL fluid ranges between 50% and 73% (15-17).
The findings from non-nasopharyngeal samples in children provide important insights into the diverse manifestations and detection of COVID-19 in pediatric patients. The significant positivity rates across various sample types highlight that while nasopharyngeal swabs are commonly used, other sample types can also play a crucial role in diagnosing COVID-19, especially in specific clinical contexts. For example, the high positivity rate in BAL samples underscores the importance of lower respiratory tract sampling in severe cases or when nasopharyngeal tests are negative despite clinical suspicion (18, 19). Similarly, the detection of the virus in CSF raises critical questions about the potential for neurological involvement in pediatric COVID-19, warranting further investigation into the mechanisms of viral entry into the CSF and its clinical implications (20). The low positivity rates in stool and sputum samples suggest that while viral shedding in the GI and upper respiratory tracts is less common, these samples can still be valuable for comprehensive diagnosis, particularly in cases presenting with GI symptoms (21). The statistically significant differences observed across sample types (P = 0.000) reinforce the importance of using a multi-faceted diagnostic approach to improve detection accuracy and patient management in pediatric populations (22).
These findings highlight the need for a broader diagnostic strategy in pediatric COVID-19 cases, particularly when standard nasopharyngeal tests are inconclusive. Future research should focus on the clinical implications of positive findings in these alternative sample types and their potential role in understanding the full spectrum of COVID-19 pathogenesis in children. This could lead to more tailored diagnostic protocols and better clinical outcomes for pediatric patients.
The hypothesis that SARS-CoV-2 can be detected in certain tissues is based on the virus’s affinity for the ACE2 receptor, which binds to the virus’s spike (S) protein. This interaction plays a key role in the virus’s ability to infect cells and is a critical determinant of both the transmission and severity of COVID-19. The widespread expression of ACE2 in various tissues may explain the virus’s presence in different clinical samples. Studies indicate that the level and affinity of the ACE2 receptor are higher in the lower respiratory tract. However, different studies have reported varying rates of infection positivity in BAL samples. It is estimated that in 11% of cases where nasopharyngeal samples test negative but there is a strong clinical or radiological suspicion, BAL samples can confirm SARS-CoV-2 infection (23).
In the present study, patients with positive BAL samples exhibited significantly higher rates of clinical symptoms such as dyspnea and cough. In a study by Wang et al., BAL samples had the highest positivity rate (93%), followed by sputum (77%), nasal swabs (63%), fibrobronchoscopy brush biopsy (46%), throat swabs (32%), stool (29%), and blood (1%). Notably, none of the 72 urine samples tested positive (13, 24, 25). In a study conducted in Italy, Turriziani et al. reported that 15% of BAL samples from suspected COVID-19 patients tested positive for SARS-CoV-2 (26). In contrast, Chang et al. in the United States examined 206 BAL samples and found no positive cases of the virus (25, 26). However, studies from China showed significantly higher positivity rates, with 93% to 100% of BAL samples testing positive for SARS-CoV-2, highlighting regional variations in detection rates (23).
Among the patients who had their ETT samples tested, 11% were positive. Statistically significant higher rates of dyspnea were reported in this group. This suggests that SARS-CoV-2 presence in the lower respiratory tract, as reflected by ETT samples, is associated with more severe respiratory symptoms like difficulty breathing, indicating a correlation between viral load in the lower respiratory system and clinical severity (27).
In the group of patients who had CSF samples taken, 17.8% tested positive, though no specific statistically significant symptoms were reported. As of now, only a small group of patients with neurological symptoms and CSF analysis have been identified as CSF RT-PCR positive. These findings suggest that viral infections were not a significant contributor to the clinical presentation. Further investigations are needed to clarify the role of SARS-CoV-2 in the development of meningoencephalitis, particularly in suspected CSF infection (28, 29).
Of the patients who provided sputum samples, 14 tested positive (2.9%), and no specific statistically significant symptoms were reported in this group either. Sputum is better than oropharyngeal sampling not only in detecting SARS-CoV-2 but also in identifying other respiratory viruses unrelated to COVID-19. This supports the use of self-collected sputum as a viable alternative to oropharyngeal sampling for COVID-19 diagnosis. Sputum provides comparable diagnostic accuracy in terms of positivity rates, sensitivity, predictive values, and viral load detection. Sputum collection also offers advantages by reducing patient discomfort and lowering healthcare workers’ exposure to infectious aerosols (30).
Gastrointestinal symptoms are increasingly being recognized as common in COVID-19 patients. Gastrointestinal manifestations include loss of appetite, nausea, vomiting, diarrhea, abdominal pain, and abnormal liver function tests. In our study, among the patients who provided stool samples, only one (4.8%) tested positive, and this patient was male. In this group, symptoms such as nausea, diarrhea, body aches, and general weakness were statistically significant. Gastrointestinal involvement and identified GI symptoms may appear in only some patients. Therefore, recognizing and understanding GI symptoms associated with COVID-19 is crucial for proper patient care.
In a systematic review and meta-analysis, it was determined that GI symptoms are frequently observed in COVID-19 patients, with diarrhea being linked to more severe illness and potentially poorer outcomes. Early identification of such patients is crucial for timely intervention and effective management of this high-risk group.
This study has several limitations that could affect the generalizability and strength of its findings. The sample sizes for certain groups, such as those with CSF and fecal samples, were relatively small, limiting statistical power. Additionally, the study was conducted in a single hospital, which may not represent broader populations. It focused on hospitalized and ICU patients, which could skew results toward more severe cases of COVID-19.
5.1. Conclusions
This study investigated the presence of SARS-CoV-2 in various clinical samples from 1,567 suspected COVID-19 patients, revealing a 12% overall positivity rate. Notably, BAL samples exhibited the highest positivity at 20.7%, indicating their effectiveness for detecting the virus in severe cases. The findings highlight significant correlations between viral presence in respiratory samples and clinical symptoms, particularly cough and dyspnea. While SARS-CoV-2 was also detected in CSF and fecal samples, the clinical implications remain unclear and warrant further investigation. The study underscores the importance of using multiple sample types for accurate diagnosis and understanding transmission dynamics, particularly regarding GI symptoms associated with COVID-19.