1. Background
Since late 2019, the global pandemic of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), which causes coronavirus disease 2019 (COVID-19), has emerged. This disease has spread rapidly across the world and infected millions worldwide (1).
The SARS-CoV-2 is typically considered a respiratory virus, with primary symptoms such as sore throat, cough, congestion, and dyspnea (1, 2). However, gastrointestinal (GI) symptoms are also recognized in some patients, who shed viral RNA in their stool both during infection and after they have apparently recovered (3).
Although SARS-CoV-2 was originally described as a viral pneumonia infection, over time it has been observed to cause multisystem disease and can affect various organs. Therefore, clinicians should be aware of potential systemic complications when treating this disease (4). Transmission of SARS-CoV-2 typically occurs through the respiratory tract, but some studies report the presence of viral particles in the stool of patients. Understanding the viable SARS-CoV-2 is crucial for comprehending this pathogen and its effects on patient health and the immune system, as well as informing public health, especially in the control of nosocomial infection committees in hospitals (1, 3). On the other hand, fecal microbiota transplantation (FMT) is gaining attention as a treatment for various conditions related to dysbiosis of the gut microbiome, such as the treatment of recurrent Clostridioides difficile infection (5, 6). The US Food and Drug Administration (FDA) issued a safety alert regarding the need for donor screening tests to identify virulent or multidrug-resistant (MDR) bacteria, such as MDR organisms, enteropathogenic Escherichia coli, and Shiga toxin-producing E. coli (6, 7).
The global spread of COVID-19 and early reports of fecal shedding of this virus have led to concerns about the possibility of SARS-CoV-2 transfer via FMT. Consequently, the FDA issued a safety alert recommending nasal testing of donors and direct testing of donor stool for the virus before FMT (6, 7).
2. Objectives
Given the importance of identifying the SARS-CoV-2 virus in various clinical samples, this study aims to track the presence of the virus genome in stool samples of children suspected of having coronavirus. The objective of this study is to identify the coronavirus in stool samples and compare it with the results obtained from nasopharyngeal samples of the same patients.
3. Methods
3.1. Study Design and Sample Collection
A total of 105 pediatric patients meeting the inclusion criteria were enrolled in this study, and informed consent was obtained. The inclusion criteria were: Age between one month and 18 years old, suspected COVID-19 patients admitted during the COVID-19 pandemic, and patients' consent to participate in the study. Demographic data, hospitalization details, clinical manifestations, SARS-CoV-2 PCR results, and ancillary laboratory findings were analyzed. Stool samples were collected from the hospital's emergency and infectious wards from suspected children with viral GI syndrome, such as fever, lymphopenia, and increased CRP and ESR during the COVID-19 pandemic. The samples were transferred to the Pediatric Infectious Research Center (PIRC) and kept at a temperature of minus 80 degrees Celsius until testing. Nasopharyngeal swabs were examined daily for the detection of SARS-CoV-2. Patients with no GI symptoms and negative preclinical data supporting GI viral infections were excluded from the study.
3.2. Stool Preparation
At the time of testing, the samples were first taken out of the freezer and brought to room temperature. Then, the stool sample was mixed with sterile DNase/RNase-free water at a ratio of 1 to 5 (weight/volume). A total of 500 µL of DNase/RNase-free water was added to the feces and thoroughly mixed and homogenized by vortexing. In the next step, the microtube was centrifuged at 5000 rpm for 5 minutes, and then 300 µL of the supernatant was taken for the extraction process.
3.3. Nasopharyngeal Swab Preparation
The nasopharyngeal swab from children suspected of having viral GI syndrome was transferred to the PIRC in viral transport media (VTM). The sample was then mixed using a vortex for 20 seconds. A total of 200 µL of the mixed VTM was transferred to a DNase/RNase-free microtube for the total RNA extraction step.
3.4. Total RNA Extraction
Total RNA from all 105 stool and nasopharyngeal samples was extracted using a commercial Total RNA extraction kit from SIMBIOLAB (Lot No.116-2326-0202N100). Some modifications were made, including the use of twice the RNA carrier concentration and the addition of proteinase K for stool samples.
3.5. Real-time PCR
Due to the difficulty of culturing viruses and the need for advanced facilities, PCR is used to identify viruses. The sensitivity and specificity of this test in identifying viral genetic material have been reported differently in various studies. However, in these studies, the sensitivity ranges from 94 to 100 percent, and the specificity ranges from 95 to 100 percent, depending on the selection of the appropriate target for primer design (8-10). Therefore, the results of the real-time PCR (COVITEC, Lot No. COVIT-21008-1 Iran) were used to evaluate the quality of the extracted RNA using a designed and commercial kit. The spike protein gene (S gene) and the nucleocapsid gene (N gene) of SARS-CoV-2, along with RNase P as an internal control, were detected using the COVITEC real-time PCR kit. The internal control was used to confirm the RNA extraction, and positive and negative controls of the COVITEC real-time PCR kit were also used to ensure the PCR process.
3.6. Statistical Analysis
Statistical analysis was conducted using SPSS software version 23. To determine the relationship of positive real-time PCR results between stool and nasopharyngeal swab samples, the Student's t-test was used. A P-value ≤ 0.05 was considered statistically significant.
4. Results
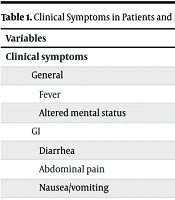
A total of 105 samples were enrolled in this study between March and September 2022. Sixty percent of participants were female. The mean age was 2.64 ± 3.44 years (ranging from 5 months to 14 years old). The duration of hospitalization was 5.98 ± 7.95 days (ranging from several hours to 30 days), and 3 (2.8%) mortality was observed. The most common symptom was fever, and all patients had diarrhea; the rest of the symptoms are presented in Table 1. Underlying diseases (such as Kawasaki disease, febrile seizures, and esophageal atresia) were observed in 11 (10.4%) patients. Laboratory hematology data such as CRP, ESR, etc., are presented in Table 1.
| Variables | Values |
|---|---|
| Clinical symptoms | |
| General | |
| Fever | 74 |
| Altered mental status | 5 |
| GI | |
| Diarrhea | 100 |
| Abdominal pain | 65 |
| Nausea/vomiting | 53 |
| Respiratory | |
| Cough | 57 |
| Sore throat | 20 |
| Hypoxemia (SpO2 < 93% air room) | 12 |
| Dyspnea | 8 |
| Preclinical data of the patients | |
| WBC (/µL) | 8.22 ± 4.59 |
| Lymphocytes (%) | 46 |
| Platelet (/µL) | 300.9 ± 122.6 |
| Hemoglobin (g/dL) | 10.83 ±1.46 |
| ESR (mm/h) | 18 ± 16.8 |
| CRP (mg/L) | 14.7 ± 12.3 |
Abbreviation: GI, gastrointestinal.
a Values are expressed as % or mean ± SD.
The RNA of SARS-CoV-2 was detected in stool samples and nasopharyngeal swabs of 37 (35%) and 35 (33%) patients, respectively, according to the results of the real-time PCR. The result patterns of real-time PCR for stool versus nasopharyngeal swab samples are shown in Table 2.
| Variables | Results | |||
|---|---|---|---|---|
| Stool samples | Positive | Negative | Negative | Positive |
| Nasopharyngeal samples | Negative | Positive | Negative | Positive |
| Number (%) | 19 (18) | 17 (16) | 51 (49) | 18 (17) |
The nasopharyngeal PCR of 17 samples (16%) from the patients was positive, while their stool PCR results were negative. The RNA of SARS-CoV-2 was detected in 19 stool samples (18%) from patients but was not detected in their nasopharyngeal samples. However, this difference was not significant in the results of stool and nasopharyngeal swab samples (P > 0.05).
5. Discussion
The lungs are the primary target of the SARS-CoV-2 virus, and the respiratory system is well known as the primary pathway for the virus to spread. The virus enters target cells by binding to the ACE-2 receptor, which is present not only in lung cells but also in cells of other organs. This raises the possibility of the virus infecting organs beyond the lungs. Hence, it is important to consider alternative transmission routes apart from the respiratory route (4, 11-14).
Widely documented fecal shedding of SARS-CoV-2 viral RNA, sometimes without positive respiratory testing, has led to an interest in assessing stool viral loads for various purposes, from diagnostics to environmental monitoring for rapid outbreak detection (7). Moreover, there is concern regarding the potential spread of SARS-CoV-2 through fecal matter, raising concerns about the risk of transmission during FMT procedures, especially considering the chance of asymptomatic individuals shedding the virus in their stool (7).
In this study, we present the frequency of SARS-CoV-2 RNA in the feces and nasopharyngeal samples of asymptomatic individuals. In the current study, SARS-CoV-2 RNA was detected in 35% of stool samples. Various studies (11, 15-17) have reported different rates of the presence of SARS-CoV-2 RNA. In studies conducted in the United Arab Emirates, Spain, and India in 2022, 53%, 43.5%, and 62% of stool samples, respectively, contained the virus (15-17).
Another study in Iran in 2022 showed that 27.03% of stool samples were positive for SARS-CoV-2 RNA (11). The frequency obtained in the current study is lower than that in the United Arab Emirates, Spain, and India (15-17) but higher than another study in Iran (11). These varying frequencies of stool-positive SARS-CoV-2 can be attributed to several factors, including the study population and variants of SARS-CoV-2.
The detection of the viral genome in various clinical samples plays a crucial role, especially in understanding its link to the severity of clinical symptoms and the mode of transmission. Therefore, this research was conducted to examine the presence of SARS-CoV-2 genetic material in stool and nasopharyngeal samples from patients suffering from COVID-19, as well as to explore how it correlates with the clinical manifestations experienced by individuals (11).
Numerous diagnostic methods have been extensively investigated to identify individuals with infections since the onset of the COVID-19 pandemic (11, 18). The results of this study indicated that SARS-CoV-2 was more frequently detected in stool samples than in nasopharyngeal samples in pediatric patients with the GI symptoms of COVID-19, but there was no significant difference between the results of the stool and nasopharyngeal PCR results (P > 0.05).
Additionally, the duration of viral shedding from stool can be measured if patient follow-up is conducted. These data can be very helpful for epidemiology. A larger sample size can better confirm the results and make them generalizable to all similar studies.
5.1. Conclusions
The findings of this study showed that stool samples can be a good alternative for detecting SARS-CoV-2 in patients with GI symptoms. The study confirmed the existence of the SARS-CoV-2 genome in the stool samples of individuals diagnosed with COVID-19, indicating the potential for transmission of the virus through the oral-fecal route. Viral RNA shedding in stool is important in FMT and environmental infection. Therefore, it is important to identify the biological sample containing the virus genome. It is crucial to prevent the shedding of the virus and the spread of infection.
5.2. Limitations
We encountered some limitations in this study, such as being single-center, having a small sample size, and lacking longitudinal follow-up. It would have been better to monitor patients over time or measure how long viral RNA persisted in stool samples, but we were unable to conduct this process.