1. Background
Treatment of infections caused by Pseudomonas aeruginosa is becoming increasingly complicated by its tendency to acquire resistance to multiple classes of antibacterials. The agents commonly used to treat these infections include Antipseudomonal, Penicillins, Aminoglycosides, Carbapenems, Fluoroquinolones, and Cephalosporins (1). The emergence of resistance to antibiotics in P.aeruginosa is a serious concern. The beta-lactamases-producers are capable of hydrolyzing almost all β-lactam antibiotics and are associated with high morbidity and mortality (2). Methylation of 16S rRNA has emerged as a mechanism of high-level Aminoglycoside resistance among bacteria in recent years, and prevention of the adverse effects of intrinsic Aminoglycosides that would block their own 16S rRNA. Aminoglycosides continue to play an important role in the management of serious infections caused by bacteria, often in combination with broad-spectrum beta-lactams. Since 2003, the production of 16S rRNA methylase has been detected to be a novel mechanism of aminoglycoside resistance. So far, seven types of methylases have been identified (ArmA, RmtA, RmtC, RmtB, RmtE, RmtD, and NpmA). Also, these genes are mediated by bacterium-specific recombination systems, such as transposons, and are easily translocated to other DNA target sites (3). AmpC beta-lactamases hydrolyze expanded-spectrum cephalosporins. The most plasmid-mediated AmpC β-lactamases belong to the DHA, FOX, and CMY families (4). In 1989, Bauernfeind et al. detected a K. pneumoniae isolate from South Korea that could transfer resistance to cefoxitin and cefotetan as well as to Penicillins, Oxyimino-cephalosporins, and Monobactams to E.coli. The enzyme, termed CMY-1 for its Cephamycinase activity, and was more sensitive to be inhibited by Sulbactam than Clavulanate or Tazobactam, suggesting that it might be a class C enzyme (5). Since 1998, three plasmid-mediated quinolone resistance (PMQR) mechanisms have been described, namely Qnr, aac(6′)-Ib-cr and QepA. The rise in multiresistant bacteria harboring aac(6')-Ib has seriously limited the effectiveness of Amikacin and other Aminoglycosides. These mechanisms are widely prevalent amongst common clinical isolates (6). The present study was conducted to determine the prevalence of these enzymes among P. aeruginosa clinical isolates from Iran.
2. Objectives
The present study was conducted to determine the prevalence of CMY, aac (6′)-Ib and 16S rRNA methylase genes among Pseudomonas aeruginosa isolates from Iran.
3. Patients and Methods
3.1. Isolation and Clinical Identification
From September to January 2011, 448 burnt patients referred to Shahid Motahari hospital (level I burn care center) in Tehran, Iran were recruited. 100 isolates of P. aeruginosa were collected by sterile swabs; before ulcers were washed by physiological serum. Samples were transferred in stuart media, cultured on Cetrimide and MacConkey agar and incubated at 37ºC for 24 h. Colonies that made pigment or odor, were studied using biochemical tests such as oxidase test, catalase, sugar fermentation test, and ability of growth in 42ºC. The isolates were stored at -200c in brain heart broth containing 20% glycerol. Pseudomonas aeruginosa ATCC27853 was used as a control strain.
3.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility to Imipenem (IPM, 10 μg), Meropenem (MEM, 10 μg), Ceftazidime (CAZ, 30 μg), Amikacin (AK, 30 μg), Tobramycin (TOB, 10 μg), Piperacillin/Tazobactam (PTZ, 100/10 μg), Ciprofloxacin (CIP, 5 μg), Cefepime (FEP, 30 μg), Ceftriaxone (CRO, 30 μg), Aztreonam (ATM, 30 μg), Gentamicin (GEN, 10 μg) and Carbenicillin (Car, 100 μg) (Mast Group, Merseyside, UK) was performed by the Kirby-Bauer disk diffusion method on Mueller Hinton agar (Merck, Germany) based on clinical laboratory standards institute (CLSI) guidelines (7). Pseudomonas aeruginosa ATCC27853 was used as a control strain.
3.3. Minimum Inhibitory Concentration
Minimum inhibitory concentration (MIC) of different antibiotics (Imipenem, Meropenem, Cefepime, Ampicillin, Piperacillin/Tazobactam, Ceftriaxone and Ceftazidime (GLAXO England Co. and Himedia Co.) were determined according to the guidelines of the CLSI by broth microdilution method (7).
3.4. Detection of Resistance Genes
DNA templates were prepared by boiling method. All clinical P.aeruginosa isolates were screened for the presence of CMY, aac(6′)-Ib, armA, rmtB, rmtC and rmtD by polymerase chain reaction (PCR) amplification in a thermal cycler (Eppendorf, Master cycler gradient). The primer pair sequences used in the study has been shown in Table 1, and genes were amplified under the following condition.
| Nucleotide Sequence | Primer | Target | PCR Conditions | Size (bp) | ||
|---|---|---|---|---|---|---|
| Denaturing | Anneal | Extension | ||||
| (5′-ATGATGAAAAAATCGTTATG-3′) (5′-TTGTAGCTTTTCAAGAATGC-3′) | CMY-F, CMY-R | CMY | 94°C, 45 s | 55°C, 45 s | 72°C,45 s | 635 |
| (5′-GCTTTCTGCGGGCGATGTAA-3′) (5′-ATGCAATGCCGCGCTCGTAT-3′) | rmtB-F, rmtB-R | rmtB | 94°C, 45 s | 55°C, 45 s | 72°C, 45 s | 173 |
| (5′-CGAAGAAGTAACAGCCAAAG-3′) (5′-ATCCCAACATCTCTCCCACT-3′) | RmtC-F, RmtC-R | rmtC | 94°C, 45 s | 55°C, 45 s | 72°C, 45 s | 711 |
| (5′-ATTCTGCCTATCCTAATTGG-3′) (5′-ACCTATACTTTATCGTCGTC-3′) | ArmA-F, ArmA-R | armA | 94°C, 45 s | 55°C, 45 s | 72°C, 45 s | 315 |
| (5′-CGGCACGCGATTGGGAAGC-3′) (5′-CGGAAACGATGCGACGAT-3′) | RmtD-F, RmtD-R | rmtD | 94°C, 30 s | 55°C, 30 s | 72°C, 30 s | 401 |
| (5′-TTGCGATGCTCTATGAGTGGCTA- 3′) (5′-CTCGAATGCCTGGCGTGTTT-3′) | aac(6′)-Ib-F, aac(6′)-Ib-R | aac(6′)-Ib | 94°C, 45 s | 55°C, 45 s | 72°C, 45 s | 482 |
Oligonucleotide Primers Used in This Study
3.5. Nucleotide Sequence Accession Number
The nucleotide sequence data reported in this paper have been submitted to the GenBank sequence database and assigned accession no.JX648311 and JX644173.
4. Results
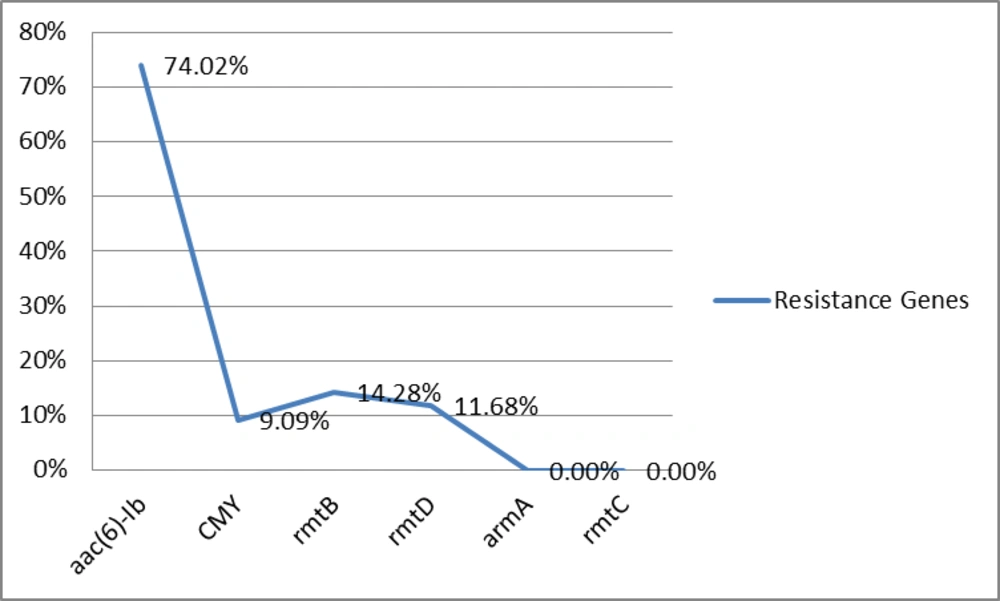
All P. aeruginosa isolates were resistant to Meropenem, Amikacin, Tobramycin, Ciprofloxacin, Aztreonam, Piperacillin/Tazobactam, Ceftriaxone, Cefepime, and Carbenicillin. Seventy-seven (77%) of 100 isolates were resistant to Imipenem and Ceftazidime, while 49% of isolates were resistant to Gentamacin. Also isolates were multidrug resistant (MDR) (resistant to more than three antibiotics from different classes were defined as MDR). aac(6)-Ib, CMY, rmtB and rmtD were detected in 57 (74.02%), 7 (9.09%), 11 (14.28%), and 9 (11.68%) isolates, respectively. No armA and rmtC genes were detected in isolates (Figure 1).
5. Discussion
Recent 20 years, P.aeruginosa is known as the most common bacteria in burn wards in Tehran, Iran (8). 16S rRNA methylases can confer high-level resistance to aminoglycosides (3).These genes have been found in Pseudomonas aeruginosa and other bacteria in many areas. The dissemination mechanisms of 16S rRNA methylases genes are of great clinical importance. In the present study, the rmtB gene was more prevalent than the arma, rmtD and rmtC genes in P.aeruginosa isolates. The rmtB and rmtD genes were found in 11 (14.28%) and 9 (11.68%) isolates, respectively and armA and rmtC were not found totally. A number of different pan-aminoglycosides resistance-promoting 16S rRNA methylases have been detected in P.aeruginosa, including RmtA, RmtB, RmtD and ArmA (9). In a study from China, armA or rmtB were detected in 94.7% (89/94) of Acinetobacter baumannii and Pseudomonas aeruginosa strains, and armA was predominant (84 vs. 5 strains with rmtB). No rmtA, rmtC, rmtD or npmA genes were found (10). In a study from France, four armA isolates with positive results were detected in 2005 (n = 2) and in 2007 (n = 2), in addition to a single isolate with positive result from 2006. No isolate had positive results for the rmtA, rmtC, rmtD, and npmA genes (11). The amp-C beta-lactamases constitute a group of enzymes widely speared in gram-negative bacteria which inactivate Cephalosporins such as Ceftazidime and Cefotaxime (4). The CMY gene was detected in 7 (9.09%) of isolates. In Taiwan, five isolates were found to produce CMY-2 AmpC enzymes in a total of 1210 isolates, and one isolate carried both CTX-M-3 and CMY-2, also three and nine isolates expressed putative AmpC β-lactamases (12). A novel aminoglycoside acetyltransferase that exhibits FQ-acetylating, aac(6')-Ib-cr, has been detected in P.aeruginosa (9). The aac (6)-Ib gene was detected in 57 (74.02%) of isolates. In a study from the United States, among 313 Enterobacteriaceae, aac(6′)-Ib was present in 50.5% of isolates (13). The prevalence of coproduction of rmtB, rmtD, aac(6)-Ib and CMY in this collection of Imipenem-resistant P. aeruginosa isolates was 3 (3.8%). This is the first report of CMY, aac(6′)-Ib and 16S rRNA methylase genes among Pseudomonas aeruginosa isolates from Iran. Therefore, control and treatment of these infections caused by the mentioned bacteria is difficult, so there is a need for revise treatment protocols to prevent resistant genes dissemination among clinical isolates.