1. Background
Extended-spectrum β-lactamases (ESBLs) have been observed in virtually all species of the family Enterobacteriaceae mainly Escherichia coli and Klebsiella pneumoniae (1). Among ESBLs the most widespread and clinically relevant are class A ESBLs of TEM, SHV, and CTX-M types. CTX-M-type β-lactamases are increasingly becoming the predominant ESBLs globally in recent years, including Asia (2). These enzymes have been classified into five major groups by amino acid sequence similarities: clusters of CTX-M-1, CTX-M-2, CTX-M-8, CTX-M-9, and CTX-M-25/26. They have a preferential hydrolysis of Cefotaxime over Ceftazidime; although some CTX-M type ESBLs, including CTX-M-15, show good activity against Ceftazidime (3). Recent advances in the molecular characterization of antibiotic resistance mechanisms have resulted in the discovery of genetic elements, called integrons which can integrate antibiotic resistance genes. Several studies have reported integron distributions in multidrug resistant strains isolated from animals and humans. The role of integrons in the development of multiple resistances relies on their unique capacity to acquire gene cassettes and express cassette-associated genes (4). An integron, which can be located either on the bacterial chromosome or on a plasmid, includes the gene for an integrase site (int) and for an adjacent recombination site (attI). So far three classes of antibiotic-resistance-encoding integrons have been identified. Each class has its own integrase. Among the antibiotic–resistance integrons, class 1 integrons are the most common integron type, class 2 integrons are embedded in Tn7-family transposons and only one example of a class 3 integron is known (5).
2. Objectives
The aim of this study was to investigate the antimicrobial susceptibility and to determine the prevalence of CTX-M-1 group enzymes and class 1, 2, and 3 integrons genes among K. pneumoniae isolated from children in Tehran hospitals, Iran.
3. Patients and Methods
3.1. Bacterial Isolates and Identification
A total of 31 nonduplicate Klebsiella pneumoniae isolates were collected from clinical specimens of children aged 0–12 years admitted to three hospitals in Tehran (Loghman-E Hakim in the southwest, Imam Khomeini in the center, and Milad in the northwest) between May and December 2011. They were identified as K. pneumoniae using biochemical tests and a PCR method to confirm the identification of K. pneumoniae subsp. pneumoniae, using published specific primer pair for K. pneumoniae 16S– 23S internal transcribed spacer gene (6): Pf: 5′-ATTTGAAGAGGTTGCAAACGAT-3′ and Pr1: 5′-TTCACTCTGAAGTTTTCTTGTGTTC-3′ (amplicon size: 130 bp). Genomic DNA was prepared by boiling the isolates and used as the templates (7). Cycling conditions were as follows: initial denaturation at 94°C for 5 min; 35 cycles of 94°C for 1 min, 58°C for 1 min, and 72°C for 1 min followed by a final elongation at 72°C for 7 min. K. pneumoniae ATCC13883 was used as positive control.
3.2. Antibiotic Susceptibility Testing
The antibiotic susceptibilities were determined by disk diffusion method on Mueller-Hinton agar plates (Merck, Darmstadt, Germany) as recommended by the clinical and laboratory standards institute (CLSI) (8). The disks containing the following antibiotics (μg) were used (Mast, UK): Cefotaxime (CTX, 30), Ceftriaxone (CRO, 30), Ceftazidime (CAZ, 30), Imipenem (IPM, 10), Amoxicillin-Clavulanic acid (AUG, 30), Aztreonam (ATM, 30), Ciprofloxacin (CIP, 5), Tobramycin (TN, 10), Tetracycline (T, 30), Trimethoprim-sulfamethoxazole (TS, 25), Gentamicin (GM, 10), Cefepime (CPM, 30), Cefoxitin (FOX, 30), and Amikacin (AK, 30). E. coli ATCC 25922 was used as quality control for antimicrobial susceptibility.
3.3. ESBL Screening and Confirmation by Phenotypic Methods
The isolates showing reduced susceptibility to Ceftazidime or Cefotaxime or both were tested for ESBLs production by the combined disk method according to the CLSI guidelines (8). The inoculum and incubation conditions were the same as for standard disk diffusion recommendations. Combined disk method was performed using four disks (μg): Cefotaxime (CTX) (30), Cefotaxime + Clavulanic acid (10), Ceftazidime (CAZ) (30), and Ceftazidime + Clavulanic acid (10). A≥ 5 mm increase in zone diameter for either antimicrobial agent tested in combined with Clavulanic acid versus its zone when tested alone was designated as ESBL positive (8). Quality control for the production of ESBLs was performed using E. coli ATCC 25922 and K. pneumoniae ATCC 700603 as negative and positive controls, respectively.
3.4. CTX-M-1 β-lactamase and Class 1, 2, and 3 Integrons Identification
As mentioned above, genomic DNA was prepared by boiling the isolates and used as the templates for PCR reactions (7). Amplification of all genes was performed on Gene Amp PCR System PTC-1148 (Bio-Rad, Foster City, CA, USA).
For detection of blaCTX-M-1 ESBL genes, PCR was performed using primers for blaCTX-M-1 group genes: 5′-GGTTAAAAAATCACTGCGTC-3′ and 5′-TTGGTGACGATTTTAGCCGC-3′ (amplicon size: 864 bp) (9). Amplification of class 1, 2, and 3 integrons was performed for blaCTX-M-1 positive isolates using published specific primer pairs for intI1, intI2, and intI3 genes as follows: 5′-CAGTGGACATAAGCCTGTTC-3′ and 5′-CCCGAGGCATAGACTGTA-3′(for intI1, amplicon size: 160 bp) (10), 5′-CACGGATATGCGACAAAAAGGT-3′ and 5′-GTAGCAAACGAGTGACGAAATG-3′(for intI2, amplicon size: 789 bp) (11), and 5′-GCCTCCGGCAGCGACTTTCAG-3′ and 5′-ACGGATCTGCCAAACCTGACT-3′(for intI3, amplicon size: 979 bp) (11). Multiplex PCR was used to detect intI2 and intI3 genes.
Cycling conditions were as follows: Initial denaturation at 94°C for 5 min; 35 cycles of 94°C for 1 min, 54°C for 1 min, and 72°C for 1 min followed by a final elongation at 72°C for 7 min. Conditions were the same for all genes except for intI2 and intI3 multiplex-PCR for which the annealing temperature was 60°C. The PCR products were analyzed by electrophoresis with 1% agarose gels in 1X TAE (Tris-Acetate-EDTA) buffer. The gels were stained with 5 μg/mL ethidium bromide, and the PCR products were visualized under the UV light.
4. Results
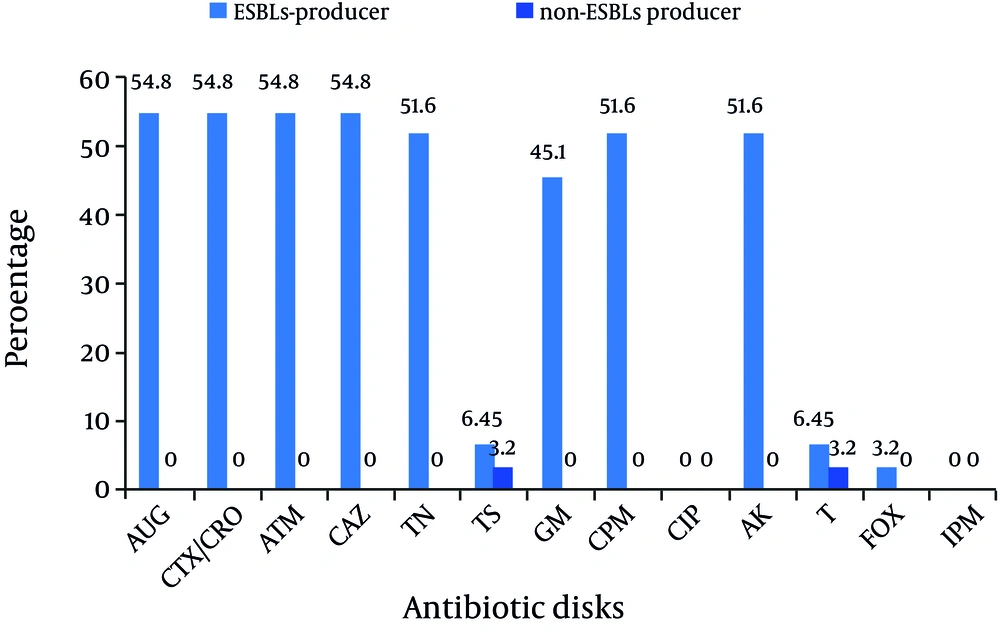
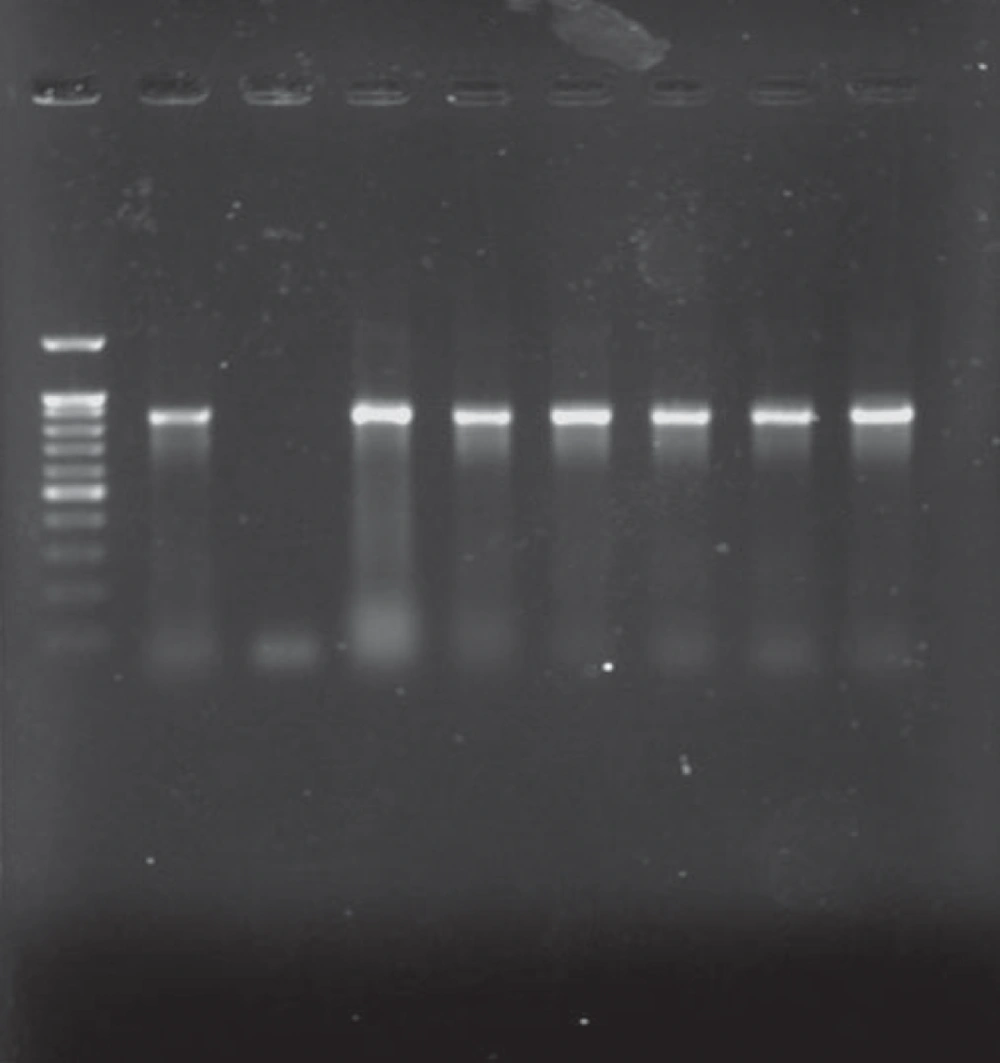
All K. pneumoniae isolates with positive findings for biochemical tests also showed positive PCR results for the 16S–23S internal transcribed spacer region. Of 31 isolates, 21 were isolated from urine (67.7%), 2 from wound, 2 from tracheal secretions, and 6 from other samples (including catheter, eye, and etc.). 24 strains were isolated from females (24/31, 77.4%). Eleven strains were isolated from NICU (35.5%), 11 from pediatric (35.5%), and 6 from Neonatal wards (19.3%) (Table 1). Analysis of the antimicrobial susceptibility profile of the isolates showed that all were susceptible to Imipenem and Ciprofloxacin. Of 31 isolates, 54.9% were simultaneously resistant to Amoxicillin-Clavulanic acid, Cefotaxime, Ceftriaxone, Aztreonam, and Ceftazidime (n = 17), 16 isolates were resistant to Tobramycin, Cefepime, and Amikacin (51.6%), 14 isolates were resistant to gentamicin (45.2%), 3 isolates were resistant to Trimethoprim-Sulfamethoxazole, and Tetracycline (9.67%), and 1 isolate was resistant to Cefoxitin (3.2%). Among 31 isolates, 17 (54.9%) were identified as ESBLs producers according to the combined disk method. ESBLs-producer isolates demonstrated a higher degree of multidrug resistance as compared to non-ESBLs (Figure 1) (P value < 0.05). The blaCTX-M-1 group gene was detected in all cefotaxime-resistant isolates (17/31, 54.9%) (Figure 2).
| Strains | Hospital | Sex | Ward | Specimen |
|---|---|---|---|---|
| 1 | Milad | Fa | NICU a | Urine |
| 2 | Milad | Ma | Pediatric | Urine |
| 3 | Milad | F | NICU | Urine |
| 4 | Milad | F | Pediatric | Urine |
| 5 | Milad | F | Neonatal | Urine |
| 6 | Milad | F | Neonatal | Urine |
| 7 | Milad | F | NICU | Urine |
| 8 | Milad | F | NICU | Urine |
| 9 | Milad | F | Pediatric | Urine |
| 10 | Loqhman | M | Pediatric | Wound |
| 11 | Milad | M | NICU | Peritoneal secretions |
| 12 | Milad | F | Pediatric | Urine |
| 13 | Milad | M | Pediatric | Urine |
| 14 | Milad | F | NICU | Eye |
| 15 | Milad | F | NICU | Urine |
| 16 | Milad | F | NICU | Tracheal secretions |
| 17 | Milad | F | Pediatric | Urine |
| 18 | Imam | M | Pediatric | Urine |
| 19 | Imam | F | NICU | Eye |
| 20 | Imam | F | Neonatal | Urine |
| 21 | Imam | F | NICU | Tracheal secretions |
| 22 | Imam | F | Neonatal | Umbilical secretions |
| 23 | Imam | F | Neonatal | Urine |
| 24 | Imam | M | Pediatric | Urine |
| 25 | Imam | F | Neonatal | Urine |
| 26 | Milad | M | PICU a | Catheter |
| 27 | Milad | F | NICU | Urine |
| 28 | Milad | F | Emergency | Urine |
| 29 | Milad | F | Pediatric | Wound |
| 30 | Loqhman | F | Pediatric | Urine |
| 31 | Milad | F | PICU | Throat secretions |
aAbbreviations: F, female; M, male; NICU, neonatal intensive care unit; PICU, pediatric intensive care unit
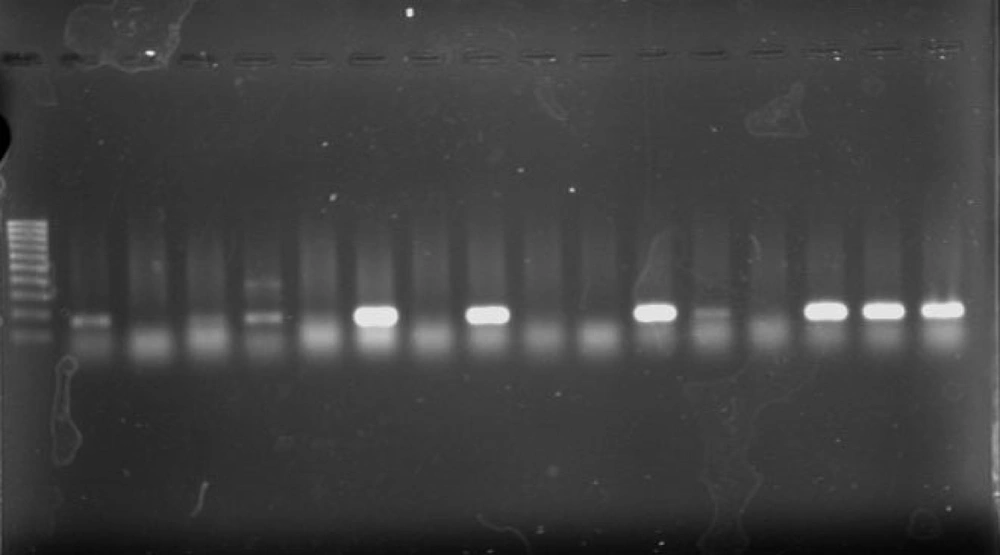
Using the PCR assay, 8 (25.8%) of 31 isolates were found to have positive results for the presence of class 1 integrons (Figure 3). Five strains were isolated from urine (5/8, 62.5%) and 7 were isolated from females (7/8, 87.5%). Two strains were isolated from NICU (2/8, 25%), 2 from neonatal (2/8, 25%), 3 from Pediatric (3/8, 37.5%), and 1 from PICU ward. No integrons were found in the remaining isolates. No class 2 or class 3 integrons were detected among isolates.
5. Discussion
Infections caused by ESBL-producing bacteria are increasing in many countries including Iran (12, 13). During the last decades, the emergence of antimicrobial drug-resistant strains has been reported in K. pneumoniae isolated from community and hospital acquired infections (14-16). Resistance to antimicrobial agents is often associated with the spread of transmissible plasmids and integrons which can be located on the chromosome or plasmids. The ability of integrons to integrate resistance gene cassettes makes them prime pools for the further dissemination of antibiotic resistance among clinical isolates of gram-negative bacteria, including K. pneumoniae (17).
In this study, the frequency of ESBL-producing K. pneumoniae isolated from children was 54.9% (17/31). The blaCTX-M-1-group gene was detected in 54.9% of isolates. In a study performed by Karimi et al., 19 isolates from 50 K. pneumoniae isolated from children with urinary tract infections (UTI) (38%), had positive results for ESBLs production (18). In another study performed by Seyed Javadi et al., Of 30 K. pneumoniae isolated from children with UTI, 21 (70%) were multidrug resistant (19). In a study performed by Feizabadi et al., the prevalence of blaCTX-M-1 genes among the K. pneumoniae isolates was 45.2% (13). Several reports have confirmed the emergence of CTX-M-1-producing K. pneumoniae isolates in Sweden, France, Madagascar, and Croatia. Also in Asian countries, CTX-M-1-producing K. pneumoniae isolates have been reported from India, Kuwait, Saudi Arabia, Malaysia, The Philippines, Singapore, and Thailand (2, 20, 21). Various factors involved in these differences including strains isolated from hospitalized patient or from outpatient, the geographic differences across the world, and etc.
In our study, 25.8% of isolates were found to havepositive results for the presence of class 1 integrons. No class 2 or class 3 integrons were detected. Karimi et al., reported that the prevalence of class 1 integrons was 48% in K. pneumoniae isolates (18) and Seyed Javadi et al., reported that class 1 integrons were found in 13.3% of the K. pneumoniae isolates (19). In Seyed Javadi study, class 2 integron was not found which is in accordance with our study. In a survey in Taiwan on the prevalence of class 1 integrons in clinical K. pneumoniae isolates collected from Taiwan, during 2 periods (1993 and 2004), class 1 integrons were present in 78 isolates (34.2%) from 1993 and 129 (32.9%) from 2004 (22). The most commonly encountered integrons are those of class 1 which may be located on transmissible plasmids and transposons (23).
In conclusion, the results of our study revealed that CTX-M-1- producing K. pneumoniae is already present in some parts of Tehran. K. pneumoniae is a very efficient hospital pathogen and, in addition, can readily acquire plasmid-mediated CTX-M-1 genes from E. coli. High rates of antimicrobial resistance and the role of integrons in the acquisition and dissemination of resistance genes among bacteria suggest to monitor mechanisms of antimicrobial resistance and to emphasize on the rational use of antimicrobials to decrease the spread of ESBL producing bacteria.