1. Background
Staphylococcus aureus (S. aureus) is one of the most important infectious agents in nosocomial infections, causing a wide variety of clinical signs ranging from mild to systemic and potentially fatal disorders either from invasion or toxin production; especially in patients with compromised immune systems or disabilities (1, 2). Methicillin resistant S. aureus (MRSA) isolates are versatile pathogens with high resistance rates to antibiotic therapy. MRSA isolates are nosocomial and community acquired and considered as a threat to healthcare settings (3). Community associated MRSA isolates can cause virulent infections and may spread more easily from person to person or cause more skin disease (due to Panton- Valentine Leukocidin toxin) (4). MRSA isolates are resistant to the beta-lactam antibiotics by producing penicillin binding protein 2a (PBP2a) with significantly reduced affinity for these antibiotics (5). PBP2a is encoded by staphylococcal cassete chromosome mec (SCCmec). Colonization and pathogenicity of S. aureus is a complex process involving a diverse array of virulence factors controlled by a network of virulence regulators (6). Quorum sensing is a process of cell-cell communication that enables bacteria to recognize cell density and response to extracellular signaling molecules called autoinducers. The many processes controlled by quorum sensing include bioluminescence, biofilm formation, antibiotic production, competence, sporulation and virulence factor production (7). Quorum sensing in S. aureus is based on the agr system that encoded by the agr locus. The accessory gene regulator (agr) is an important gene regulator by encoding the specific peptide called auto-inducing peptide (AIP). As an important quorum sensing system, this gene regulator is a chromosomal locus comprising of two transcripts called RNAII and RNAIII. RNAII operon (or P2 promoter) which is sensitive to AIP concentration, encodes agr A, B, C and D. AIP is encoded by agr D gene, then post-translationally modified or processed by agr B product. However, agr C component encodes a histidine kinase (agr C), a sensor for auto inducer peptide and activates agr A. The response regulator that activates P2 or P3 promoters is the agr A component. Regulation of either of these promoters subsequently leads to RNAII or RNAIII transcription (8). The adhesive surface proteins are regulated positively by RNAIII transcript during logarithmic phase of growth. There are suggestions about the correlation between agr groups and the kind of infection (3, 4).
2. Objectives
This study was conducted to detect the agr specificity groups among methicillin resistant and susceptible children S. aureus isolates.
3. Materials and Methods
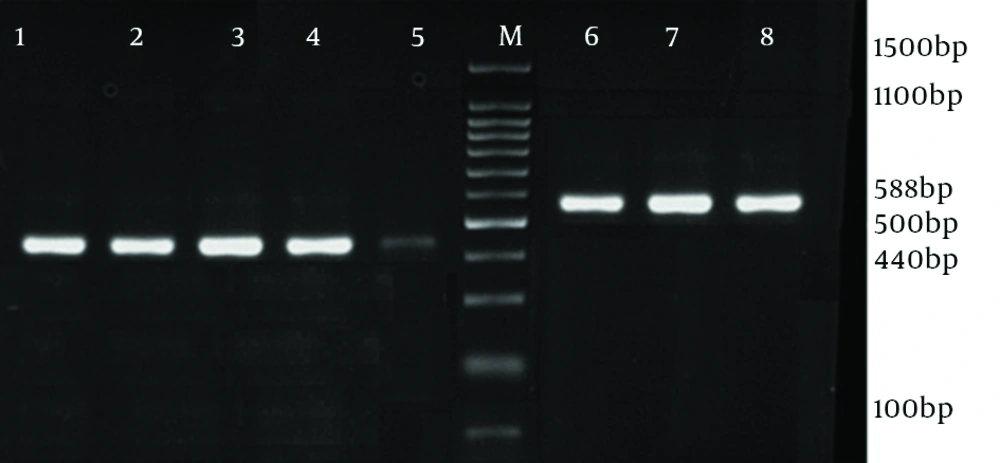
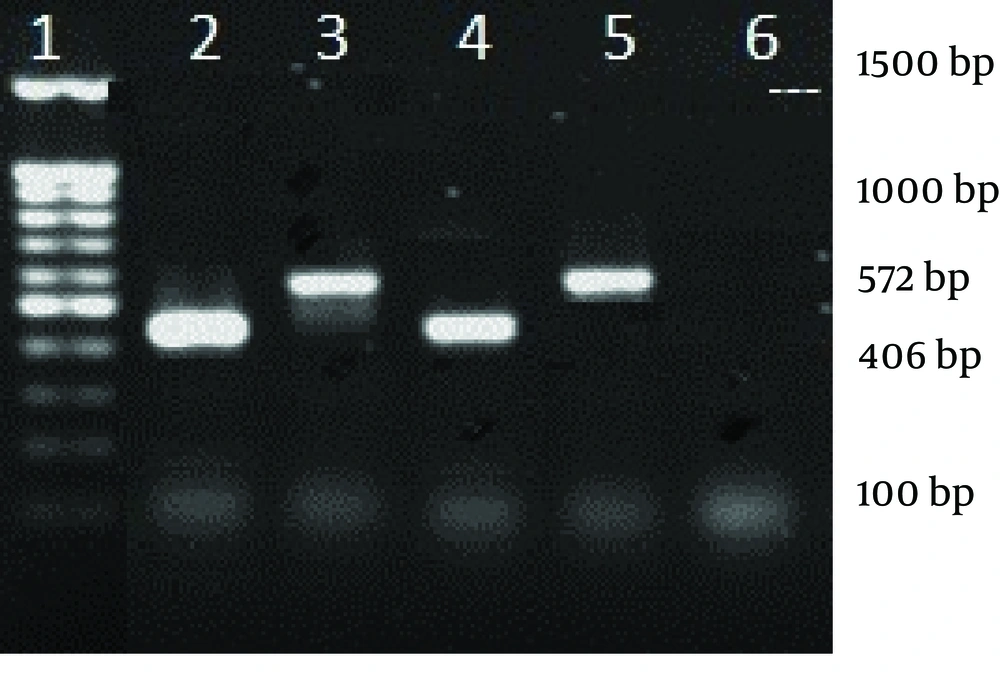
From July 2012 to January 2013, a total number of 22 S. aureus clinical isolates were collected from trachea (14 isolates), blood cultures (4 isolates), skin lesions (2 isolates) and sputum (2 isolates) from hospitalized children (13 male and 9 female) with different clinical signs, aged between 2 to 11 years, in Loghman Hospital of Tehran. The identification tests were conducted by conventional tests (catalase, slide and tube coagulases, growth on mannitol salt agar and DNase tests). Antimicrobial susceptibility test was performed by Kirby Bauer assay (disk diffusion method), according to clinical and laboratory standards institute (CLSI) guidelines. S. aureus ATCC 25923 was used as control strain in this study. The discs used in this study included: oxacillin (1 µg), tetracycline (30 µg), clindamycin (2 µg), erythromycin (15 µg), vancomycin (2 µg), linezolid (30 µg), trimethoprim-sulfamethoxazole (10 µg), amoxicillin (10 µg), gentamycin (120 µg) and ciprofloxacin (5 µg) (from MAST, UK). DNA extraction was performed by boiling method (TE buffer [Tris-Hcl + EDTA, pH 8] and lysostaphin [2 µg/mL, 20 µL]) (9), and then the DNA was stored at -20°C. PCR assay was performed to detect mecA gene and agr specificity groups (by duplex PCR). Primers for mecA and agr specificity groups has been shown in Table 1 (3, 10, 11).
| Primer | Sequence 5′3′ | Size (bp) |
|---|---|---|
| mecA | ||
| F: GTG AAG ATA TAC CAA GTG ATT | 147 | |
| R: ATG CGC TATAGATTGAAA GGA | ||
| agrI | ||
| F: ATGCACATGGTGCACATGC | 440 | |
| R: GTCACAAGTACTATAAGCTGCGAT | ||
| agrII | ||
| F: ATGCACATGGTGCACATGC | 572 | |
| R: GTATTACTAATTGAAAAGTGCCATAGC | ||
| agrIII | ||
| F: ATGCACATGGTGCACATGC | 406 | |
| R: CTGTTGAAAAAGTCAACTAAAAGCTC | ||
| agrIV | ||
| F: ATGCACATGGTGCACATGC | 588 | |
| R: CGATAATGCCGTAATAC CCG |
3.1. Data Analysis
Data were analyzed by Pearson Chi-Square. A P value less than 0.05 was considered as statistically significant.
4. Results
The antibiotic susceptibility pattern was as follows: resistance to amoxicillin was observed in 14 isolates (63.6%), tetracycline in eight (36.3%), ciprofloxacin in five (22.7%), gentamycin in four (18%), trimethoprim-sulfamethoxazole in four (18%), erythromycin in six (27.2%) and clindamycin in four (18%) isolates. All o isolates were susceptible to vancomycin and linezolid. MRSA isolates were detected with oxacillin disk (1 µg) during disk diffusion and then confirmed by detection of mecA gene. In 12 (54.5%) isolates which had 440 bp in electrophoresis, agr group I was found. Among these, 10 isolates (55.6%) were MSSA and two (50% of MRSA isolates) were MRSA. Three MSSA isolates (16.7%) and one MRSA (25%) belonged to agr group II. In three isolates (11.11% of MSSA and 25% of MRSA) agr specificity group III was detected, while agr specificity group IV was detected in four (18%) isolates, involving two MSSA (11.7%) and one MRSA (25%). Table 2 shows the characteristics of five MRSA isolates all were resistant to at least 6 antibiotics of which two resistant to the all, except for linezolid and vancomycin. Four isolates belonged to agr specific group I and one isolate to agr group IV. Prevalence of agr specific groups in MRSA and MSSA is presented in Table 3. There was no statistical significant difference between methicillin resistant and susceptible clinical isolates regarding presence of agr groups (P = 0.13). Furthermore, there was no relationship between infection types and agr groups (Figures 1 and 2).
| Isolates (MRSA) | Clinical Sample | Sex | agr Groups | Antibiotic Resistance |
|---|---|---|---|---|
| 1 | Blood | M | I | T, A, CD, E, CIP, GM |
| 2 | Lesion | F | IV | T, A, CD, E, CIP, GM |
| 3 | Trachea | M | I | T, A, CD, SXT, E, CIP, GM |
| 4 | Lesion | M | I | T, A, CD, SXT, E, CIP, GM |
| 5 | Trachea | M | I | T, A, CD, SXT, E, CIP |
aAbbreviations: A, amoxicillin; CD, clindamycin; CIP, ciprofloxacin; E, erythromycin; GM, gentamycin; SXT, trimethoprim- sulfamethoxazole; T, tetracycline.
| Isolates | agr I | agr II | agr III | agr IV |
|---|---|---|---|---|
| MSSA (n = 17) | 10 (55.6) | 3 (16.7) | 2 (11.11) | 2 (12) |
| MRSA (n = 5) | 2 (40) | 1 (20) | 1 (20) | 1 (20) |
| Total (n = 22) | 12 (54.5) | 4 (18) | 3 (9) | 3 (18) |
a Data are presented as No. (%).
5. Discussion
Community acquired methicillin resistant S. aureus (CA-MRSA) can affect healthy children with high morbidity and mortality (12). In the present study, all isolates were susceptible to vancomycin and linezolid and had high resistance to amoxicillin, suggesting the presence of plasmids encoding beta-lactamases, easily transmitted to other strains. There was higher antibiotic resistance in MRSA isolates, such as aminoglycosides and tetracycline. This is especially common in hospital associated isolates (13). These findings were also confirmed by Adebayo’s study, in which all isolates were susceptible to vancomycin and linezolid, and also antibiotic resistance was more prevalent in MRSA strains (14). Armin reported resistance to vancomycin and linezolid (15). Quorum sensing system regulates staphylococcal virulence factors. This system participates in staphylococcal scaled skin syndrome (agr specificity groups III), exfoliative toxin production (agr group IV), and encoding surface proteins (16-18). Also, agr group I was the most prevalent specific group (54.5%), due to present study’s findings, confirming previous surveys (19). In the Ho CM study, 91.6% of the clinical isolates had agr group I (20). Also in the Barbara study, 45.7% of the clinical isolates were agrI positive (21). This group has been detected mostly in suppurated infections such as endocarditis (3). The second prevalent groups were agr groups II and IV (18% and 13.6%, respectively). Group II agr has been more associated to respiratory infections, especially in community acquired MRSA (3, 22). The rate of group III prevalence was 9% which has been shown to be prevalent in isolates with active vancomycin resistance mechanisms (7). Quorum sensing system leads Staphylococcus aureus to express virulence factors (23), alter the antibiotic susceptibility pattern, resist the immune system and form biofilm. It was shown that agr group I is the most frequent specific group among children, also the most prevalent group in MSSA and MRSA isolates and there was no statistically significant difference between two groups, while MRSA strains had significantly higher antibiotic resistance.