1. Background
Staphylococcus aureus (S. aureus) colonizes nearly 25 - 30% of skin or nose of healthy people. The methicillin-resistant S. aureus (MRSA) is a type of Staphylococcus that is resistant to certain antibiotics, such as methicillin, cloxacillin, dicloxacillin, oxacillin, nafcillin, and closely related class of drugs, such as cephalosporins (e.g. cephalexin). Broad-spectrum antibiotic overuse for less severe infections is one of the most important reasons of MRSA expansion.
Unfortunately, these MRSA isolates, which are susceptible only to glycopeptides antibiotics, such as vancomycin, are becoming multidrug resistant (1). At present, low level resistance to vancomycin is appearing and increasing (2).
The possible predisposing factors of MRSA emergence are: long duration of hospitalization, consumption of antibiotics without medical prescription, lack of awareness, receipt of antibiotics before coming to the hospital, etc. (3).
The MRSA serious infections have been increasing throughout the world. Infected patients and health care providers carriers play an important role in spreading and transferring this superbug in the hospital (4). Today, the emergence of multiple drug resistance and monitoring of disease transmission by MRSA isolates, in hospitals and also in communities, represent the major challenges of the healthcare systems (5).
In approximately 1 - 2% of people, MRSA is detected on their skin or in their nose. Infections due to MRSA are not different from any other Staphylococci infections, although several strains of MRSA may be more virulent than regular Staphylococci. Identification of infection due to MRSA requires laboratory testing, especially the antibiogram test, because without knowing the antibiotic resistance pattern, it is more difficult to manage MRSA infections.
Unfortunately, penicillin resistant strains spread in healthcare facilities and in the community. Methicillin narrow spectrum semi-synthetic penicillin was introduced to overcome infections caused by beta-lactamase-producing S. aureus. First MRSA strain was identified in 1961. It was isolated from the hospital environment and named as hospital acquired S. aureus (HASA) (6).
1.1. Staphylococcal Cassette Chromosome mec Typing
Staphylococcal Cassette Chromosome mec (SCCmec) typing, which classifies SCCmec elements on the basis of their structural differences, is applied in epidemiological studies to distinguish MRSA strains. A MRSA clone defines the genotype of a methicillin-sensible S. aureus (MSSA) strain with a combination of SCCmec elements (6).
Two essential components in the SCCmec elements of Staphylococci are the ccr gene complex (ccr) and the mec gene complex (mec). The ccr gene complex contains ccr genes, open reading frames (ORFs) and the mec gene elements.
In conformity to the following classification, several allotypes have been found among SCCmec elements:
a) Type I: class B mec + type 1 ccr; b) type II: class A mec + type 2 ccr; c) type III: class A mec + type 3 ccr; d) type IV: class B mec + type 2 ccr; e) type V: class C2 mec + type 5 ccr.
The presence of the mecA causes resistance of S. aureus to methicillin. The penicillin binding protein 2a (PBP2a) has a low affinity for all beta-lactam antibiotics compared to other PBP. In the presence of a beta-lactam antibiotic, the peptidoglycan layer with PBP2a is not disrupted and the bacterium can survive. The mecA gene and its regulatory genes locate within a mec operon together: mecI and mecR1. It is proposed that the mec operon in S. aureus was achieved from S. sciuri and the mecA-positive coagulase negative Staphylococci (CoNS), particularly S. epidermidis. Also, there is the acquirement of the mecA region from S. fleurettii, a commensal bacterium of animals (6).
2. Objectives
With regard to the above topics, we decide to determine the prevalence of MRSA in health care providers, their antimicrobial resistance pattern and the SCCmec typing.
3. Materials and Methods
3.1. Microbiological Methods
In this descriptive study, two hundred and twenty nine health care providers, such as nurses and health care workers, were examined from different wards in Mofid Children Hospital, Tehran, Iran. Office personnel were excluded. Nasal samples for examination of S. aureus and sociodemographic data were obtained from persons during one year, from August 2012 to July 2013. All subjects in this study had no underlying diseases and have not taken antibiotics 2 weeks before sampling. Specimens were taken from the health care providers as follows: a sterile moistened swab was inserted into each nostril to approximately 1 cm depth, and rotated five times. The samples were transferred quickly to the laboratory and were inoculated onto mannitol salt agar medium and incubated at 35°C for overnight. The isolates were identified as S. aureus based on morphologic and biochemical tests such as: Gram stain, catalase, coagulase, hot-cold β-hemolysin on blood agar, DNAase and mannitol salt agar fermentation (7). All the strains were screened for methicillin resistance by oxacillin (1 µg) and cefoxitin (30 µg) disk diffusion test, based on the standard guidelines (8).
3.2. Antibiotic Resistant Pattern
The resistant patterns of MSSA and MRSA strains were determined by disk diffusion method (Kirby-Bauer). The antibiotics panel was: penicillin (10 units), cefpodoxime (10 µg), oxacillin (1 µg), vancomycin (30 µg), linezolid (30 µg), clindamycin (2 µg), ciprofloxacin, rifampicin (5 µg), teicoplanine (30 µg), cefepime, erythromycin (15 µg), cefotaxim (30 µg), azithromycine (15 µg), and ceftazidim (30 µg), minocycline (30 µg), doxycycline (30 µg), trimethoprime-sulfametoxazol (25 µg), ceftriaxone (30 µg). Zone diameters, as recommended by the clinical and laboratory standards institute (CLSI 2012), were measured after 24 hours incubation at 35°C. American type culture collection (ATCC) 29213 S. aureus was used, as the control strain (8).
3.3. Staphylococcal Cassette Chromosome mec Typing
The DNA was extracted from all MRSA isolates with the AccuPrep genomic DNA extraction kit (cat.no.K-3032 lot no.1008J, BIONEER, Seoul, Korea). The polymerase chain reaction (PCR) components and amplification profiles comprise: 300 nM concentration of each oligonucleotide primer (Eurofins MWG Operon LLC, Huntsville, AL, USA); 5.5 mM MgCl2; 200 mM each deoxynucleoside triphosphates (dATP, dCTP, dGTP, and dUTP); and 0.125 U of Taq DNA polymerase (prime Taq TM DNA polymerase, lot no: 100914, cat.no. A type: G-1002, URL GENET BIO, Daejeon, Korea).
Multiplex PCR for SCCmec typing was performed, based on Oliveira and Lencastre methods. The seven different loci (loci A to H) along the mecA gene and mecA gene cassette were selected for amplification by PCR (Table 1) (9).
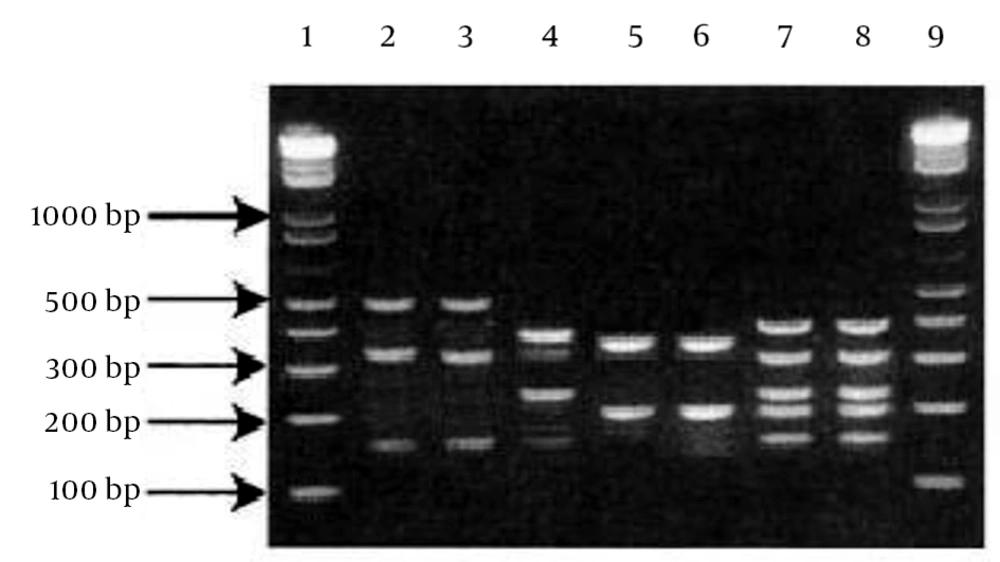
The PCR products were analyzed by gel electrophoresis on 1.2% Bioneer agarose gels (cat.no.c-9100-1 lot.no.1101c, Bioneer, Seoul, Korea) in 1x TBE buffer (890 mM Tris, 890 mM of boric acid, 40 mL of 0.5 M EDTA, pH 8.0) at 100 V, for 65 minutes. Green loading buffer with DNA stain (lot: 111.034, Jena Bioscience, Jena, Germany) was used when loading the samples and ladder. The sizes of the PCR products were determined by comparison with the molecular size standard (50 bp - 1 Kb linear scale; low range DNA ladder or 100 bp - 3 Kb linear scale and mid-range DNA ladder, Jena Bioscience) (8, 10-12).
3.4. Statistical Analysis
Statistical analysis was conducted using the SPSS version 16. Fisher’s exact test was used to evaluate the relation between MRSA and MSSA. A P <0.05 was considered as significant.
| Gene | Primer Sequences | Size, bp |
|---|---|---|
| Type I, specific locus A | 495 | |
| Forward | 5´-TTCGAGTTGCTGATGAAGAAGG-3´ | |
| Reverse | 5´-ATTTACCACAAGGACTACCAGC-3´ | |
| Type II, specific locus B | 284 | |
| Forward | 5´-ATTCATCTGCCATTGGTGATGC-3´ | |
| Reverse | 5´-CGAATGAAGT GAAAGAAAGTGG-3´ | |
| Type II, III, specific locus C | 209 | |
| Forward | 5´-ATCA AGACTTGCATTCAGGC-3´ | |
| Reverse | 5´-GCGGTTTCAATTCACTTGTC-3´ | |
| Type I, II, IV, specific locus D | 342 | |
| Forward | 5´-CATCCTATGATAGCTTGG TC-3´ | |
| Reverse | 5´-CTAAATCATAGCCATGACCG-3´ | |
| Type III, specific locus E | 243 | |
| Forward | 5´-GTGATTGTTCGAGATATGTGG-3´ | |
| Reverse | 5´-CGCTTTATCT GTATCTATCGC-3´ | |
| Type III, specific locus F | 414 | |
| Forward | 5´-TTCTTAAGTA CACGCTGAATCG-3´ | |
| Reverse | 5´-GTCACAGTAATTCCATCAATGC-3´ | |
| Nonspecific locus G | 381 | |
| Forward | 5´-CAGGTCTCTTCAGATCTACG-3´ | |
| Reverse | 5´- GAGCCATAAACACCAATAGCC-3´ | |
| Nonspecific locus H | 303 | |
| Forward | 5´-C AGGTCTCTTCAGATCTACG-3´ | |
| Reverse | 5´-GAAGAATGGGGAAAGCTTCAC-3´ | |
| Specific mecA | 162 | |
| Forward | 5´-TCCAGATTACAACTTCACCAGG-3´ | |
| Reverse | 5´-CCACTTCATATCTTGTAACG-3v |
Primer Sequences of Staphylococcal Cassette Chromosome mec Typing Genes
4. Results
In this study, 229 health care providers (23 - 49 years old) from 16 different hospital wards (infectious, gastrointestinal, pediatric intensive care unit, neonatal intensive care unit, endoscopy, neonatal, hematology, neurology, surgery, nephrology, respiratory, dialysis, emergency, laboratory, radiology and pediatric infectious research center) were studied. Two hundred (87.33%) were female and 29 (12.66%) were male. No significant differences were observed in MRSA colonization between health care providers in various wards. Staphylococci were isolated in 27 cases (12%). A total of 21 (77.7%) cases were MRSA while six (22.3%) cases were MSSA. No significant difference was seen between the age (P = 0.920), gender (P = 0.315) and type of ward in MRSA and MSSA carriers.
The antibiotic resistance pattern in MRSA was more expanded than in MSSA, although the difference was not significant. The MSSA was sensitive to most antibiotics. All strains in this study were sensitive to linezolid and vancomycin, and the rate of penicillin resistance was high in both groups (Table 2).
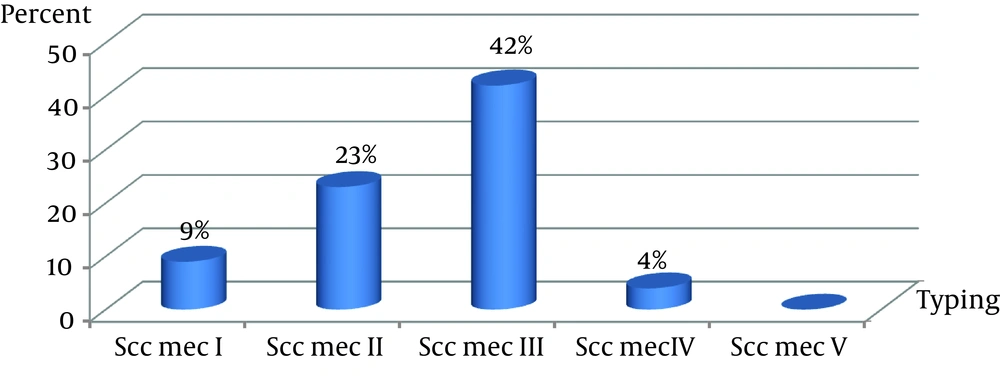
All MRSA isolates had the mecA gene. From 21 MRSA, only four S. aureus strains were non-typable by the primers used in this study. The SCCmec type III group (42%) and SCCmec type II (23%) were the two most common types among the health care providers. The lowest incidence was in the SCCmec type IV (Figure 1). The PCR products of SCCmec were analyzed by gel electrophoresis on 1.2% agarose gel (Figure 2).
| Antibiotics | MRSA/Resistant, N = 21 | MSSA/Resistant, N = 6 | P Value |
|---|---|---|---|
| Azithromycine | 10 (47.62) | 0 | 0.057 |
| Erythromycin | 11 (52.38) | 0 | 0.054 |
| Clindamycin | 11 (52.38) | 0 | 0.054 |
| Penicillin | 20 (95.24) | 5 (83.33) | 0.402 |
| Trimethoprim/Sulfametoxazol | 4 (19.4) | 0 | 0.545 |
| Doxycycline | 6 (28.57) | 0 | 0.284 |
| Minocycline | 3 (14.28) | 0 | > 0.999 |
| Teicoplanine | 6 (28.57) | 2 (33.33) | > 0.999 |
| Rifampicin | 5 (23.8) | 0 | 0.555 |
| Cefpodoxime | 12 (57.14) | 2 (33.33) | 0.385 |
| Ceftazidim | 17 (80.95) | 4 (66.67) | 0.588 |
| Cefotaxim | 4 (19.4) | 2 (33.33) | 0.588 |
| Ceftriaxone | 12 (57.14) | 2 (33.33) | 0.385 |
5. Discussion
The worldwide emergence of MRSA is a remarkable challenge for public health (16 - 18) Based on centers for disease control (CDC) reports, 1% of all Staphylococcal infections and 50% of healthcare-associated Staphylococcal infections are caused by MRSA (3).
In this examination of 229 samples, 21 (12%) MRSA strains were detected. Similarly with our results, in trials conducted in Germany, in 2007, (13) and west of Iran, in 2013 (14), the prevalence of MRSA isolates among health care providers carriers was reported 11.3% and 17.57%, respectively.
Compared to studies in Germany (6.5%), The Netherlands (1.4%), Shiraz, Iran (5.3%), Pediatric Infections Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran (3.2%), Switzerland (3.3%), the USA (3.4%), France (6.6%) and the UK (6.7%), the prevalence of MRSA strains were lower than in our study (9, 15-21).
Rezaei et al. considered colonization with methicillin resistant and methicillin sensitive Staphylococcus aureus subtypes in patients with atopic dermatitis. They found a higher rate (33%) of MRSA colonization in the nasal cavity. The MRSA was one of the most frequent organisms that was found on their skin (22).
The high percentage of MRSA in health care providers, especially who do not exhibit any symptoms or signs of severe disease, is very dangerous, mainly because they can cause epidemic diseases, raise the occurrence of severe diseases among patients, and enhance mortality rate by transfer of the strains to patients (22).
Linezolid is one of the most effective oral medications used for outpatient treatment of MRSA infections that are resistant to other antibiotics. In this study, there was no resistance against linezolid, in both groups (23).
Resistances to antibiotics among the MRSA isolates were higher compared to MSSA, although without no significant difference was between them. The MRSA isolates showed variable resistance to clindamycin, ceftriaxone, cefpodoxime, azithromycine, and erythromycin (23). Resistance to penicillin and clindamycin (23, 24) was similar with the other studies. Moderate resistance to other conventional antibiotics (such as azithromycin, erythromycin, clindamycin, cefpodoxim, ceftriaxone was detected in MRSA (22).
By definition, all MRSA isolates carry the mecA gene, which confers resistance to all beta-lactam antibiotics, including cephalosporins and carbapenems. In our study and in similar studies, several MRSA are susceptible to a number of beta lactams, such as cephalosporins (25-27).
Several additional auxiliary factors increase MRSA susceptibility to beta-lactams or other clinically used antibiotics. These auxiliary genes, including femX (fmhB), murE, pbp2, SAV1220, SAV175 and femD (glmM), were regularly identified to give back beta-lactam susceptibility of MRSA strain context. Supplementary essential genes contain the majority of the mur and pbp genes and involve synthesizing the peptidoglycan precursor that shares this phenotype by antisense incorporation, although to different extents among several beta-lactam antibiotic classes. Genes not formerly known to change MRSA beta-lactam susceptibility, including those involved in cell division (ftsA, ftsW, ftsZ), transcription (hu), secretion (spsB), cell wall teichoic acid biosynthesis (tarL) and SAV1892, are susceptible to interfere in cell wall synthesis or repairing (28).
In every region, rating of the resistance or sensitivity of MRSA against conventionally antibiotics is different. When antimicrobials are considered for therapy, susceptibility testing for antibiotics for every isolate of MRSA should be done. This study showed that all MRSA isolates were significantly less sensitive to antibiotics, compared with MSSA isolates (24).
A remarkable result in this study was the high percentage of MRSA in health care providers. Unfortunately, it is considered that the rate of MRSA in health care providers carriers is lower than in clinical samples. Therefore, MRSA screening in these persons is not often performed in Iran. The best program for the monitoring of MRSA spread and infection remains to debate formally. However, studies have consistently indicated that screening is advantageous in high-risk units, to discover the reservoir and to begin contact cautions. Therefore, management programs may be useful to decrease MRSA infection in health care providers (29). Current studies reveal an important change of carrier rate, ranging from 0% to 29% (23-25, 30-34).
One of the most important molecular methods available for knowing the epidemiology and clonal strain relatedness of MRSA is SCCmec typing.
The SCCmec, a 21- to 67-kb mobile genetic element, is very important, because it harbors the methicillin resistance (mecA) gene and other antibiotic resistance markers that can transfer these characteristics to other bacteria, such as Enterococci. If MRSA propagates, it will become a major nosocomial pathogen worldwide.
Usually, SCCmec types I‒IV are prevalent among healthcare providers’ isolates, whereas the type IV SCCmec is the dominant element in community associated MRSA (CA-MRSA) (7, 18). Although we detected SCCmec types I‒IV, no SCCmec type V was identified.
There is a significant relation between the SCCmec types I, II and III and the hospital-acquired MRSA (HA-MRSA). However, the SCCmec type IV in S. aureus isolates has several different genetic backgrounds. It is smaller than the other SCCmec types, because it does not carry any additional resistance genes, which may facilitate its mobility. The SCCmec type IV carries functional recombinases and it has been found in multiple clones, even in the HA-MRSA, which is more mobile than the other types (9, 35-37).
As we determined the SCCmec types of isolates, we identified the SCCmec type IV element in our isolates. Usually, HA-MRSA strains carry SCCmec types I, II, and III, while the type IV element is generally carried by CA-MRSA (9, 37-39). Eventually, strains with several types of SCCmec have resulted from the genetic context, with additional virulence characteristics and the ability to spread (9, 40).
With attention to control infection, isolates harboring this SCCmec element should be treated as MRSA. Likewise, they are determined to be mecA negative by PCR. Therefore, when isolates with unclarified resistance phenotypes are detected, they require further characterization and should be transferred to specialized laboratories, until updated routine assays are accessible (39, 41).
Conventional methods for MRSA screening need to be reconsidered and only using phenotypic approaches for detection should be abandoned. Given the high rates of MRSA in health care providers in this study, detecting the carriers and decolonizing them, to reduce transmission of S. aureus in the hospital, is important. Annual screening of these persons, along with patients, is recommended.